
Quantitative proteomic analysis of urinary exosomes in kidney stone patients
Introduction
Nephrolithiasis is one of the most common diseases in urology. Approximately 10% to 12% of the general population will suffer at least one kidney stone in their lifetime (1). The prevalence of kidney stones appears to be increasing in Western societies over the past four decades (2). Moreover, the recurrence rate for kidney stones after the first treatment is reported to be as high as 50% in 10 years (3). Randall’s plaque is an ectopic calcification in the interstitial tissue of the renal papilla. It consists of calcium phosphate and may serve as the nidus for a kidney stone (4). However, mechanism underlying the plaque formation remains uncertain. Transmission electron microscopy (TEM) shows that Randall’s plaque is surrounded by calcified membrane vesicles and collagen fibers, indicating that the plaque may nucleate via membrane vesicles and grows by addition of crystals at its periphery (5).
Exosomes are important vesicles for substance transport and information transmission between cells. Specific biomolecules (such as proteins, RNA, and DNA) can be encapsulated into exosomes and secreted into the extracellular milieu. Secreted exosomes could enter and regulate the physiological function of target cells, which leads to following pathophysiological changes (6). It has been reported that exosomes are involved in many diseases, such as immune reactions, tumor metastasis, neurodegeneration, and infection (7). Few studies have demonstrated the role of exosomes in stone formation. Jayachandran et al. report that the number of urinary CD63-positive (one of exosome markers) vesicles is greater in kidney stone patients than in healthy controls (8). In addition, oxalate could promote renal tubular epithelial cells to secret exosomes (9). However, how these increased exosomes contribute to kidney stone is unclear. Singhto et al. reveal that exosomes from calcium oxalate-treated macrophages could enhance proinflammatory cytokine production and show greater binding capacity to crystals, which has been proved to be induced by proteins in exosomes (10).
Recently, urinary exosomes have attracted much attention because they can be collected noninvasively and exosomal proteins can act as biomarkers for certain disease-related pathophysiological events. Proteomic analysis of urinary exosomes has been conducted in several urinary diseases, such as bladder cancer, prostate cancer, and kidney diseases (11). While, there has been few studies evaluating the proteomics of urinary exosomes from kidney stone patients. In the present study, liquid chromatography-tandem mass spectrometry (LC-MS/MS) label-free quantitative proteomics was conducted to compare protein profiles in urinary exosomes between kidney stone patients and healthy controls. We aim to identify dysregulated proteins in urinary exosomes from kidney stone patients and to explore the potential role of exosomal proteins in nephrolithiasis.
Methods
Participants and urine collection
Patients who came to our department for surgical treatment with kidney stones were invited to participate in the study. Those with urinary tract infection, hematuria or other organic diseases were excluded. Stone composition was analyzed postoperatively for every patient. People who came to our hospital for regular physical examination were invited to participate as healthy controls. Those with urinary stones, urinary tract infection, hematuria, or other systemic diseases were excluded. Each participant was asked to provide 200 to 300 mL first morning urine in a clean collection bag for exosome and protein extraction. The urine samples were stored at 4 °C and processed within 6 h. The study was conformed to the provisions of the Declaration of Helsinki (as revised in 2013). Ethical approval was obtained from the Institutional Review Board of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (2019S1147). Informed consent was obtained from the subjects.
Exosome isolation and purification
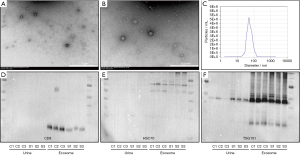
Urine exosomes were isolated and purified using modified ultracentrifugation according to previous studies (12,13). Firstly, 150 mL urine was centrifuged at 2,000 g and 4 °C for 30 min to remove cells, debris, bacteria, and the majority of Tamm-Horsfall protein (THP). Next, the remaining macropolymers and THP were removed by further centrifugation at 17,000 g and 4 °C for 60 min. The supernatant was then concentrated to 25 mL by centrifugation in 100 kD ultrafiltration centrifuge tubes (Millipore, USA) at 3,000 g and 4 °C. The concentrated urine sample was then centrifuged at 200,000 g and 4 °C for 60 min. After that, the pellet was resuspended in 15 mL phosphate-buffered saline (PBS) and centrifuged again at 200,000 g and 4 °C for 60 min. Finally, the supernatant was discarded and the pellet (exosomes) was stored at –80 °C until use. The size distribution of exosomes was evaluated by nanoparticle tracking analysis (NTA) using ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany). The morphology of exosomes was assessed by TEM using a Tecnai 12 G2 transmission electron microscope (FEI, Eindhoven, Netherlands).
Sample preparation for LC-MS/MS analysis
Exosomes were resuspended in 100 µL PBS and lysed in 200 µL 1.5× ice-cold lysis buffer [1% sodium deoxycholate (SDC), 100 mM Tris-HCl (Ph =8.5), 10 mM Tris (2-chloroethyl) phosphate and 40 mM chloroacetamide] for 5 min. Afterward, the samples were boiled for 10 min and centrifuged at 12,000 g and 4 °C for 15 min to obtain the supernatant. Protein concentration was measured using the Bradford method (Beyotime Institute of Biotechnology, China). An equal amount of protein from each sample was used and diluted with double distilled H2O to reduce the SDC concentration to <0.5%. Trypsin was added at a ratio of 1:50 (enzyme: protein) and the mixture was digested overnight at 37 °C. The next day, an equal volume of 1% formic acid in ethyl acetate was added to stop the digestion. The digest was subjected to peptide purification using self-made styrene divinylbenzene-reversed phase sulfonate desalting columns. The peptide eluate was dried under vacuum and stored at −20 °C for later use.
LC-MS/MS analysis
LC-MS/MS analysis was carried out in a hybrid quadrupole-time-of-flight (TOF) LC-MS/MS instrument (TripleTOF 5600, SCIEX) equipped with a nanospray source; 2 µg of peptides was dissolved in MS loading buffer (0.1% formic acid), loaded onto a C18 trap column (5 µm, 5 mm × 0.3 mm, Agilent Technologies) through an auto-sampler, and then eluted into a C18 analytical column (75 µm × 150 mm, 3 µm particle size, 100 Å pore size, Eksigent). Mobile phase A [3% dimethyl sulfoxide (DMSO), 97% H2O, 0.1% formic acid] and mobile phase B (3% DMSO, 97% acetonitrile, 0.1% formic acid) were used to establish an 100 min gradient, as follows: 0 min in 5% B, 65 min of 5–23% B, 20 min of 23–52% B, 1 min of 52–80% B, 80% B for 4 min, 0.1 min of 80–5% B, and a final step of 5% B for 9.9 min. The flow rate was constant at 300 nL/min. For information dependent acquisition mode analysis, each scan cycle consisted of one full-scan mass spectrum (with m/z ranging from 350 to 1,500, ion accumulation time 250 ms), followed by 40 MS/MS events (m/z ranging from 100 to 1,500, ion accumulation time 50 ms). The threshold for MS/MS acquisition activation was set to 120 cps for +2 to +5 precursors. Former target ion exclusion was set at 18 s.
Protein extraction
Urine protein was extracted using acetone precipitation. Briefly, fresh urine was centrifuged at 1,500 rpm and 4 °C for 15 min. After that, the supernatant was mixed with ice-cold acetone at ratio of 1:1. The mixture was stored overnight at −20 °C and then centrifuged at 12,000 g and 4 °C for 10 min. The pellet was dissolved in RIPA buffer (Beyotime Institute of Biotechnology, China) supplemented with phenylmethanesulfonyl fluoride (Beyotime Institute of Biotechnology, China). Briefly, the protein in purified exosomes was also extracted with RIPA buffer containing phenylmethanesulfonyl fluoride.
Western blot analysis
The concentration of extracted protein was determined using the bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, China). Equivalent amount of protein was separated using 10% or 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After being transferred to 0.2 µm polyvinylidene fluoride membranes, protein bands were blocked with 5% bovine serum albumin for 0.5 h and then incubated with primary antibody: rabbit anti-CD9 antibody (Abcam/ab92726, USA), mouse anti-Hsc70 antibody (Abcam/ab2788), rabbit anti-TSG101 antibody (Abcam/ab125011), rabbit anti-S100A8 antibody (Abcam/ab92331), rabbit anti-S100A9 antibody (Abcam/ab92507), or rabbit anti-S100A12 antibody (Abcam/ab37657) at 4 °C overnight. After that, membranes were incubated with secondary antibodies (Boster, China) conjugated with horseradish peroxidase for 1 h at room temperature and then visualized with a chemiluminescence imaging system (ChemiDoc™ MP Imaging System, Bio-Rad, USA).
Data analysis
The raw data from the TripleTOF 5600 were analyzed with MaxQuant software (V1.6.2.10) using the Andromeda database search algorithm and the MaxLFQ function. Spectrum files were searched against the UniProt human protein database using the default parameters except for the followings: label-free quantification mode was used, the min ratio count was set to 1, and the “match between runs” function was checked. The search results were filtered using a 1% false discovery rate (FDR) (at both the protein and peptide levels) and were imported into the R version 3.6.1 environment for statistical analysis. Enrichment analysis of gene ontology (GO) terms including biological process, cellular component, and molecular function was performed via DAVID 6.8 (https://david.ncifcrf.gov/) and results were filtered using a hypergeometric test with FDR <0.05.
Results
Isolation and purification of urinary exosomes
Twelve participants including six healthy controls and six kidney stone patients were involved in this study. Detailed information of participants was listed in Table 1. Primary components of all stones were calcium oxalate. Exosomes were isolated from morning urine via ultracentrifugation. TEM demonstrated that isolated samples consisted of vesicles with typical exosomal membrane morphology. NTA indicated that most vesicles showed a diameter of 60 to 90 nm and a distribution peak at 80 nm. Exosome markers (CD9, HSC70, and TSG101) were confirmed to be expressed highly in isolated vesicles by Western blotting. In addition, the expression of CD9, HSC70, and TSG101 in isolated vesicles was much higher than in the urine, which further confirmed the successful isolation and purification of urinary exosomes (Figure 1).

Full table
Proteomic profiling of human urinary exosomes
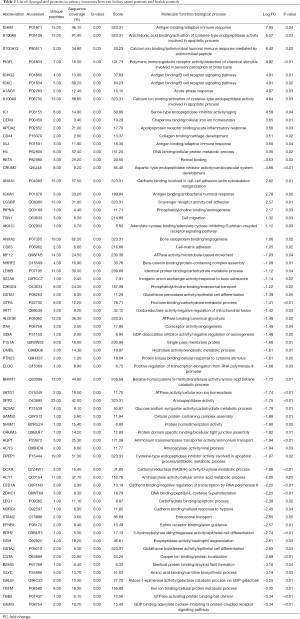
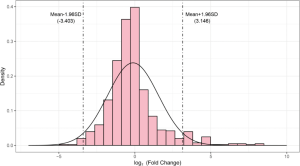
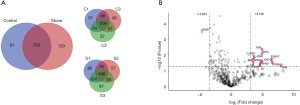
Urinary exosomal proteins from three healthy controls (C1, C2, and C3) and three kidney stone patients (S1, S2, and S3) were used for LC-MS/MS analysis. A total of 960 proteins were identified, of which 831 were identified in the control group and 879 in the stone group. Six hundred and ninety proteins were quantified at least twice in three replicates of each group and went to subsequent statistical analysis. When using absolute log2 fold change (log2FC) >1 and P<0.05 as the screening criteria, 67 dysregulated proteins were identified between two groups (Table 2). In order to identify proteins with more significant difference, we fit log2FC values to a normal distribution and detected 5% thresholds for boundary as −3.403 and 3.164 (Figure S1). Accordingly to the screening criteria (P<0.05, log2FC >3.164 or log2FC <−3.403), we identified 16 most significantly dysregulated proteins. Among these dysregulated proteins, 14 were up-regulated and two were down-regulated in urinary exosomes from kidney stone patients (Figure 2).
Full table
Bioinformatics analysis of dysregulated proteins
To explore the possible biological functions of dysregulated proteins in urinary exosomes, we conducted GO analysis using the online DAVID database. Results demonstrated that the most enriched GO terms (FDR <0.05) are related to cellular component (blood microparticle, extracellular exosome, extracellular region, and extracellular space), biological process (innate immune response, defense response to bacterium, and retina homeostasis), and molecular function (immunoglobulin receptor binding and RAGE receptor binding). Given that kidney stones are closely associated with calcium ions, we focused on another cluster named calcium ion binding (FDR >0.05 but P<0.05). S100A8, S100A9, and S100A12 proteins were found to be common in innate immune response, defense response to bacterium, and calcium ion binding. They were subsequently chosen for further study (Figure 3).

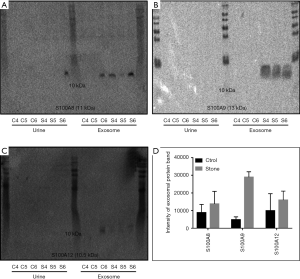
Western blotting confirmed that the expression of S100 proteins is higher in urinary exosomes from kidney stone patients
In order to validate the results obtained from LC-MS/MS analysis, western blotting was conducted to determine the level of urinary exosomal S100A8, S100A9, and S100A12 proteins in other three healthy controls (C4, C5, and C6) and three kidney stone patients (S4, S5, and S6). As shown in Figure 4, S100A8 and S100A9 were confirmed to be expressed higher in urinary exosomes from kidney stone patients than from healthy controls. Despite its low abundance in urinary exosomes, the expression of S100A12 was also higher in urinary exosomes from kidney stone patients. Detection of these three S100 proteins in urine was difficult, indicating their specific aggregation in urinary exosomes.
Discussion
Kidney stone formation involves not only urinary crystal supersaturation but also the interaction between crystals, proteins, membrane vesicles, collagen fibers, and cells. Electron microscopic examination of the renal papillary tissue at stone attachment sites has detected many membrane vesicles containing nucleated calcium phosphate crystals (14). Increased urinary exosomal vesicles are reported to be associated with kidney stones. Shyong et al. also demonstrate that calcium phosphate particles stimulate the secretion of exosomes from cultured cells (15). The pathophysiological function of exosomes is mainly dependent on the substances they contain, including proteins, RNA and DNA. Urinary exosomes contain cell-specific proteins from every segment of the nephron, which is a potential source of valuable urinary biomarkers for diseases of the kidney and urinary tract (16). We hypothesize that not only concentration, but also the protein profiles are different between urinary exosomes from kidney stone patients and healthy people. To the best of our knowledge, the present study represents the first attempt to use proteomic technology to compare urinary exosomal protein profiles between kidney stone patients and healthy controls.
Despite the fact that we included participants without urinary tract infection and hematuria, the function of dysregulated proteins in urinary exosomes from kidney stone patients focused on immune responses and defense responses to bacteria. Similarly, previous studies have also demonstrated that inflammatory processes play an important role in kidney stone formation and protein components of inflammation contribute to the abundant stone matrix proteome (17). Interestingly, we also found that some dysregulated inflammatory proteins played a role in calcium binding, namely calgranulin proteins (S100A8, S100A9, and S100A12). They were confirmed to be enriched in the urinary exosomes but not urine, suggesting that urinary exosomal S100 proteins may provide potential biomarkers for nephrolithiasis.
S100A8, S100A9, and S100A12 are members of the S100 calcium-binding protein family, which is characterized as a helix-loop-helix motif, charged amino acid residues and high affinity for calcium ion. Their expression is abundant in neutrophils and inducible in macrophages. The S100 calcium-binding protein family plays a prominent role in regulating inflammation and immune responses, including recruitment of leukocytes, production of cytokines and promotion of leukocyte adhesion and migration (18). In addition, studies have demonstrated that S100 proteins play a role in the calcification of atherosclerotic plaques. New et al. found that matrix vesicles released from calcium phosphate stimulated macrophages are rich in S100A9 and show high calcification and aggregation potential (19). Chellan et al. report that S100A12 transgenic mice could develop vascular calcification spontaneously. Overexpression of S100A12 promotes osteoblast-like cell transformation and calcification in vascular smooth muscle cells (20). Similarities between vascular calcification plaque and Randall’s plaque have been reported and osteoblast-like cell transformation of renal tubular epithelial cells is also observed during stone formation (21).
Renal tubular and collecting duct epithelium cells have also been reported to express S100 proteins (22). Unlike some classic proteins involved in nephrolithiasis [such as osteopontin (OPN) and CD44], the role of S100 proteins in stone formation is rarely studied in basic research. Instead, almost all studies focusing on protein profiles of kidney stones report that S100 proteins exist in the stone matrix (23-26). These calgranulin proteins are not only abundant in calcium oxalate stones, but also in uric acid, magnesium ammonium phosphate and matrix stones. In addition, S100A8 and S100A9 have even detected in demineralized hydroxyapatite, brushite, uric acid, calcium oxalate monohydrate, and calcium oxalate dihydrate urinary crystals precipitated from healthy human urine samples (27). More importantly, S100 proteins are restricted to the inner core but not the outer matrix of kidney stones (28), indicating that they contribute to the initial nucleation of crystals. Boonla et al. also report that S100A8 is overproduced by infiltrated leukocytes in the kidneys from nephrolithiasis patients (29). These findings highlight the relevance of S100 proteins in inflammatory pathogenesis of urolithiasis and indicate that S100 proteins are involved in the nucleation of kidney stone.
There are several limitations in our study. Firstly, the number of participants is small and studies with more participants in the future are needed to validate the current findings. Secondly, the average age of participants seems young and primary component of stones from every patient was calcium oxalate, which show a selection bias and limit the applicability to calcium oxalate stones. Thirdly, this is a descriptive proteomic study. More basic research is needed to evaluate the role of exosomal S100 proteins in stone formation.
Conclusions
Urinary exosomes from kidney stone patients are rich in S100 proteins and mainly play a role in innate immune response, defense response to bacterium and calcium-binding.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant number 81974092 and 81570631).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-41
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-41). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conformed to the provisions of the Declaration of Helsinki (as revised in 2013). Ethical approval was obtained from the Institutional Review Board of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (2019S1147). Informed consent was obtained from the subjects.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Coe FL, Evan A, Worcester E. Kidney stone disease. J Clin Invest 2005;115:2598-608. [Crossref] [PubMed]
- Scales CD Jr, Smith AC, Hanley JM, et al. Prevalence of kidney stones in the United States. Eur Urol 2012;62:160-5. [Crossref] [PubMed]
- Uribarri J, Oh MS, Carroll HJ. The first kidney stone. Ann Intern Med 1989;111:1006-9. [Crossref] [PubMed]
- Daudon M, Bazin D, Letavernier E. Randall's plaque as the origin of calcium oxalate kidney stones. Urolithiasis 2015;43:5-11. [Crossref] [PubMed]
- Khan SR, Rodriguez DE, Gower LB, et al. Association of Randall plaque with collagen fibers and membrane vesicles. J Urol 2012;187:1094-100. [Crossref] [PubMed]
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569-79. [Crossref] [PubMed]
- Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol 2015;77:13-27. [Crossref] [PubMed]
- Jayachandran M, Lugo G, Heiling H, et al. Extracellular vesicles in urine of women with but not without kidney stones manifest patterns similar to men: a case control study. Biol Sex Differ 2015;6:2. [Crossref] [PubMed]
- He Z, Guan X, Liu Y, et al. Alteration of exosomes secreted from renal tubular epithelial cells exposed to high-concentration oxalate. Oncotarget 2017;8:92635-42. [Crossref] [PubMed]
- Singhto N, Kanlaya R, Nilnumkhum A, et al. Roles of macrophage exosomes in immune response to calcium oxalate monohydrate crystals. Front Immunol 2018;9:316. [Crossref] [PubMed]
- Moon PG, You S, Lee JE, et al. Urinary exosomes and proteomics. Mass Spectrom Rev 2011;30:1185-202. [Crossref] [PubMed]
- Gonzales PA, Pisitkun T, Hoffert JD, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 2009;20:363-79. [Crossref] [PubMed]
- He L, Zhu D, Wang J, et al. A highly efficient method for isolating urinary exosomes. Int J Mol Med 2019;43:83-90. [PubMed]
- Khan SR, Canales BK. Unified theory on the pathogenesis of Randall's plaques and plugs. Urolithiasis 2015;43:109-23. [Crossref] [PubMed]
- Shyong YJ, Chang KC, Lin FH. Calcium phosphate particles stimulate exosome secretion from phagocytes for the enhancement of drug delivery. Colloids Surf B Biointerfaces 2018;171:391-7. [Crossref] [PubMed]
- Merchant ML, Rood IM, Deegens JKJ, et al. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat Rev Nephrol 2017;13:731-49. [Crossref] [PubMed]
- Sun AY, Hinck B, Cohen BR, et al. Inflammatory cytokines in the papillary tips and urine of nephrolithiasis patients. J Endourol 2018;32:236-44. [Crossref] [PubMed]
- Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer 2015;15:96-109. [Crossref] [PubMed]
- New SE, Goettsch C, Aikawa M, et al. Macrophage-derived matrix vesicles: an alternative novel mechanism for micro calcification in atherosclerotic plaques. Circ Res 2013;113:72-7. [Crossref] [PubMed]
- Chellan B, Sutton NR, Hofmann Bowman MA. S100/RAGE-mediated inflammation and modified cholesterol lipoproteins as mediators of osteoblastic differentiation of vascular smooth muscle cells. Front Cardiovasc Med 2018;5:163. [Crossref] [PubMed]
- Joshi S, Clapp WL, Wang W, et al. Osteogenic changes in kidneys of hyperoxaluric rats. Biochim Biophys Acta 2015;1852:2000-12. [Crossref] [PubMed]
- Pillay SN, Asplin JR, Coe FL. Evidence that calgranulin is produced by kidney cells and is an inhibitor of calcium oxalate crystallization. Am J Physiol 1998;275:F255-61. [PubMed]
- Okumura N, Tsujihata M, Momohara C, et al. Diversity in protein profiles of individual calcium oxalate kidney stones. PLoS One 2013;8:e68624. [Crossref] [PubMed]
- Wesson JA, Kolbach-Mandel AM, Hoffmann BR, et al. Selective protein enrichment in calcium oxalate stone matrix: a window to pathogenesis? Urolithiasis 2019;47:521-32. [Crossref] [PubMed]
- Canales BK, Anderson L, Higgins L, et al. Proteome of human calcium kidney stones. Urology 2010;76:1017.e13-20. [Crossref] [PubMed]
- Martelli C, Marzano V, Iavarone F, et al. Characterization of the protein components of matrix stones sheds light on S100-A8 and S100-A9 relevance in the inflammatory pathogenesis of these rare renal calculi. J Urol 2016;196:911-8. [Crossref] [PubMed]
- Thurgood LA, Ryall RL. Proteomic analysis of proteins selectively associated with hydroxyapatite, brushite, and uric acid crystals precipitated from human urine. J Proteome Res 2010;9:5402-12. [Crossref] [PubMed]
- Mushtaq S, Siddiqui AA, Naqvi ZA, et al. Identification of myeloperoxidase, alpha-defensin and calgranulin in calcium oxalate renal stones. Clin Chim Acta 2007;384:41-7. [Crossref] [PubMed]
- Boonla C, Tosukhowong P, Spittau B, et al. Inflammatory and fibrotic proteins proteomically identified as key protein constituents in urine and stone matrix of patients with kidney calculi. Clin Chim Acta 2014;429:81-9. [Crossref] [PubMed]


