
Comprehensive analysis of competing endogenous RNA network in Wilms tumor based on the TARGET database
Introduction
Wilms tumor (WT), also called nephroblastoma, accounted for 6–7% of malignant tumors of children under 15 years old (1). About 75% of WT were distributed in children under 5 years old (2,3). Factors that were related to abnormal genes such as tumor predisposition syndromes (4) and epigenetic mechanisms (5) might lead to WT. Recently, multiple combinations of therapy-surgery, radiation therapy or chemotherapy constitute the mainstream strategies for WT, but limited improvements have been achieved for prognosis of WT in recent years (6,7). Additionally, the prognosis of high-risk WT consisted of anaplastic WT, rhabdoid tumor or metastatic renal sarcomas were still poor (8,9). Due to the limited knowledge of its molecular pathogenesis, there was still a lack of individualized targeted management. Therefore, the molecular mechanism of WT required further exploration.
With the rapid development of high-throughput sequencing technique, we have already characterized numerous significant bio-markers across several malignancies. RNA, especially non-coding RNA has attracted more attention in recent years as it might act as a potential biomarker and drug targets (10,11). Competing endogenous RNA (ceRNA), which was proposed by Salmena et al. described a complex post-transcriptional network in which ceRNA was identified as miRNA sponges to suppress miRNA functions by competing miRNA response elements (MREs) (12). Through competition, ceRNA could cross-regulate the stability of other types of RNA and induce many genetic metabolic diseases such as tumor. LncRNA has been proved closely related to the molecular mechanism of tumorigenesis, and studies have shown that lncRNA may play an important role as ceRNA (13-16). However, few studies have explored the influence of ceRNA in WT and few results involved in prognosis have been reported.
Recently, the computational biology tools are widely utilized to develop and help generate hypotheses about the underlying mechanisms in tumor biology. Therapeutically Applicable Research to Generate Effective Treatments (TARGET) database has published a batch of information on children suffering from WT, which could be used to research the occurrence, epigenetic, transcription and other molecular mechanisms of WT. In this study, we obtained the expression profiles of lncRNAs, miRNAs, mRNAs with TARGET database. Then we constructed WT-related ceRNA network by using weighted gene co-expression network analysis (WGCNA) and Molecular Complex Detection (MCODE). What’s more, functional enrichment analysis and survival analysis was performed to reveal mechanisms and prognosis from the differentially expressed genes. This study might provide new drug biological targets for further studying to predict the prognosis of the disease. To our knowledge, this is the first study attempting to disclose the ceRNA network of lncRNAs, miRNAs and mRNAs in WT.
Methods
Population data acquisition
RNA sequencing and clinical data was obtained from the TARGET database portal (https://ocg.cancer.gov/programs/target/projects/kidney-tumors). Filtering primary data with inclusion criteria, a total of 6 non-tumor controls and 127 WT patients’ RNA-Seq data and clinical data was acquired. Of note, the control non-tumor tissues were extracted from the precancerous lesions. RNA sequence data was derived from IlluminaHiSeq RNA-Seq platforms. This study followed the publication guidelines provided by TARGET. The detailed clinical information of 127 WT patients (92 cases ≥3 years old, 35 cases <3 years old, 74 cases of female, 53 cases of male) was shown in Table 1. In terms of racial categories, Caucasian was the main component. As for tumor stage, patients with stage II [49] and III [46] were the main part in comparison with other tumor stages.

Full table
Identification of differentially expressed RNA
Raw differentially expressed data was encoded and identified based on the annotation from the Ensebml database (http://www.ensembl.org/index.html). Consequences of the differentially expression of lncRNA (DElncRNA), miRNA (DEmiRNA) and mRNA (DEmRNA) between WT tissue and normal tissue were carried out by “edgeR” package in R software (17). |log2 fold-change| >2 and the P value of false discovery rate (FDR) <0.05 were settled as a threshold (18). Besides, we generated volcano maps and heat maps of differentially expressed RNA with using heatmap package and gplots in R software.
Construction of the gene co-expression network
WGCNA was applied for obtaining the pairs of lncRNA-miRNA and miRNA-mRNA and seeking clusters of highly correlated genes (19). For the purpose of identifying the reciprocities among lncRNA-miRNA-mRNA, WGCNA was used to construct the network with a power cut-off threshold of 6 and a module size of 20. Utilizing the interaction between lncRNA-miRNA and miRNA-mRNA, gene co-expression network was constructed and visualized by Cytoscape 3.4.0. (http://www.cytoscape.org/) (20). Then, MCODE (21), a Cytoscape plugin which detected serried connected region in network extracted the hub subnetwork. Finally, Pearson’s correlation coefficient was calculated between lncRNA and mRNA in hub subnetwork.
Function annotation
Gene Ontology (GO) enrichment analysis was conducted by DAVID (database for annotation, visualization, and integrated discovery) with FDR <0.01. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis which was employed to excavate the biological action mechanisms of DEmRNAs involved in co-expression network was performed using the R clusterProfiler package (22) with the threshold of P value <0.05. Then, we applied GO plot package of R software to explore the GO terms and KEGG pathways that are closely related to tumorigenesis.
Survival analysis
In order to evaluate the prognostic value of DElncRNAs, DEmiRNAs and DEmRNAs, we performed survival analysis of those RNAs by applying “survival” package in R software. The long-rank test was employed to compare the difference. Additionally, Kaplan-Meier method was performed to generate the survival curves.
Statistical analysis
The normalization procedure was conducted via edgeR package, and Kaplan-Meier curves with log-rank test were performed by survival package. The Student’s t-test was used for continuous variables, while categorical variables were compared with Chi-square (χ2) test. Wilcoxon rank-sum test was utilized to compare ranked data with two category and Kruskal-Wallis test was utilized for comparisons among three or more groups. All statistical analysis was conducted in R studio (Version 3.5.1), and we regarded the statistical significance with P<0.05.
Results
Differentially expressed gene profiles
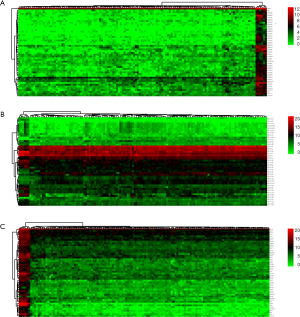
By applying the “edgeR” package in R, 127 WT tissues and 6 normal tissues were identified. A total of 5,801 differentially expressed genes were found between them, including 2,037 DElncRNAs (1,247 up-regulated and 790 downregulated), 154 DEmiRNAs (105 up-regulated and 49 downregulated), and 3,610 DEmRNAs (1,895 up-regulated and 1,715 downregulated) with thresholds of |log2 fold-change| >2 and adjusted P value <0.05. Then the heatmap and volcano map were drawn as shown in Figures 1,2.

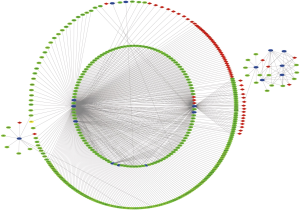
Construction of gene co-expression network
To explore how lncRNA regulated mRNA by combining miRNA in WT, pairs of lncRNA-miRNA and miRNA-mRNA were used to build ceRNA network by WGCNA. A total of 536 pairs of interacting lncRNAs and miRNAs and 4,151 pairs of interacting miRNAs and mRNAs were confirmed. Finally, 146 DElncRNAs, 62 DEmiRNAs, 287 DEmRNAs were involved in ceRNA network (Figure 3).
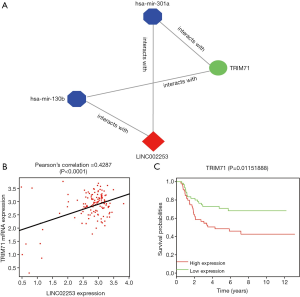
Hub subnetwork analysis
As various interacting pathways were found, we performed MCODE [21], a Cytoscape plugin to find densely connected regions in ceRNA network. As demonstrated, LINC002253 (lncRNA) was interacted with TRIM71 (mRNA) mediated by hsa-mir-301a and hsa-mir-301b (miRNA). The P value of Pearson’s correlation coefficient between LINC002253 and TRIM71 was <0.0001. Survival analysis which was utilized to identify the correlation between TRIM71 expression and clinical prognosis implied TRIM71 was negatively associated with overall survival (OS) of WT patients (the P value was <0.05) (Figure 4).
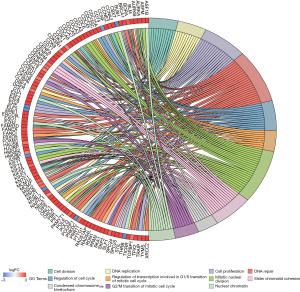
Function annotation of DEmRNA in ceRNA network
Furthermore, GO and KEGG analysis was conducted to obtain the biological functions of DEmRNAs. The GO network of biological process (BP) was shown in Figure 5, which pointed out that the most enriched GO terms in BP was tumor-related “mitotic nuclear division”. As for KEGG pathway analysis, 8 enriched pathways (Table 2) were detected. “Cell cycle”, “DNA replication”, “Oocyte meiosis” and “Homologous recombination” were considered as tumor-related pathways (Figure 6).

Full table
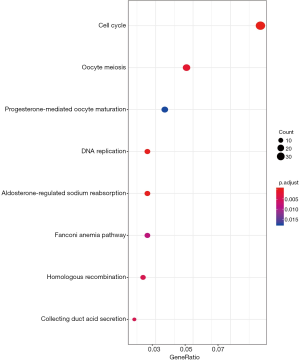
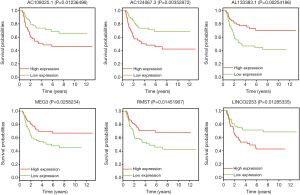
Survival analysis
The 146 DElncRNAs in ceRNA network were further analyzed by “survival” package in R software to investigate their correlations with clinical features. Kaplan-Meier curve analysis was performed to generate the survival curves. Above all, 13 DElncRNAs were significantly associated with the progression of WT. Among them, 8 lncRNAs (AC096721.1, AC108860.2, AC145422.1, AL133383.1, AL390719.2, DLGAP1-AS1, MEG3 and RMST) were associated with WT patients’ prolonged survival time, while the remaining 5 (AC108025.1, AC124067.3, AP001347.1, DLEU2 and LINC02253) were negatively correlated with OS. The top 6 most significant associated DElncRNAs were enumerated in Figure 7.
Discussion
WT is the most common neoplasm of the kidney in infants. Up to 25% of the patients suffered from chronic sequelae of disease in 25 years after being diagnosed (23). Although many researches have explored the molecular pathogenesis of WT, most of them focused on one term of lncRNA, miRNA or mRNA. As results, there was a lack of the regulatory network of overall interaction. At present, the ceRNA hypothesis, a new gene regulation term in which lncRNA could be served as the miRNA sponges to take part in regulating mRNA expression levels may be a new direction. In this study, we attempted to construct ceRNA network by using WGCNA method and explore the molecular mechanism of WT.
With more and more advances in RNA sequencing technologies, some studies suggested more than 90% of transcriptions were lncRNA so that it might be a promising prognostic biomarker (24-26). A large number of lncRNAs have been proved to be abnormally expressed in a variety of tumors, and regulating gene expression at the level of epigenetic, transcriptional and post-transcription (27-30). Sun et al. pointed out that lncRNA X-inactive specific transcript (XIST) which was down-regulated in renal cell carcinoma (RCC) tissues can up-regulate the expression of P21 through bonding with miR-106b-5p, and act as a tumor suppressor role in RCC (30).
The TARGET program applied a comprehensive genomic approach to determine molecular changes that drive childhood cancers (https://www.ocg.cancer.gov/). Such program aimed to use data to guide the development of effective, less toxic therapies. With open access to the resources of TARGET database related to WT, we confirmed the correlation among lncRNAs, miRNAs and mRNAXs and conducted a gene co-expression ceRNA network by WGCNA. One hundred and forty-six DElncRNAs were involved in WT ceRNA network. Among them, 13 (AC096721.1, AC108860.2, AC145422.1, AL133383.1, AL390719.2, DLGAP1-AS1, MEG3, RMST, AC108025.1, AC124067.3, AP001347.1, DLEU2 and LINC02253) were proved to have impact on OS of WT. Some DElncRNAs has been proved to own the potential to be biomarkers of WT. MEG3 transcripts a 1.6-kb lncRNA and has been found to inhibit the progression of several tumor cells, such as liver, colorectal, gastric, endometrial and osteosarcoma cancer cells by tumor suppressor genes p53 and Rb (31). A study suggested that MEG3 promoted metastasis and invasion of endometrial cancer cell by acting on upstream modulators or downstream effectors of p53 signaling pathway (32). In glioma tissue, upregulation of MEG3 expression significantly inhibited the proliferation of cancer in human glioma cell lines by enhancing p53 expression. What’s more, DLEU2, which was located in chromosome band 13q14, was observed to be deleted or epigenetically suppressed in leukemia, and was regarded as a negative regulator of cell proliferation. A Cytogenetic study on chronic lymphocytic leukemia patients has found that DLEU2 deletion will lead to chromosome breakage, telomere deletion, polykaryotic cells and other genetic instability (33). RMST suppressed the development of triple-negative breast cancer through inhibiting cell proliferation, invasion and migration (34). As far as we know, there was no related literature about function of AC096721.1, AC108860.2, AC145422.1, AL133383.1, AL390719.2, DLGAP1-AS1, RMST, AC108025.1, AC124067.3, AP001347.1 and LINC02253 in tumor, which deserved further exploration.
Furthermore, among all DEmRNAs in ceRNA network, TRIM71 regulated by LINC002253 was identified to be the hub gene by MCODE. TRIM71 belongs to Tripartite motif (TRIM) family proteins, most of which hold E3 ubiquitin ligase activity and can regulate intracellular signal transduction, cell apoptosis and affects tumorigenesis (35). TRIM71 was biased expressed in testis and kidney, encoding an E3 ubiquitin-protein ligase that binds with miRNAs. This gene maintained the growth of embryonic stem cells and accelerated the transformation from G1 phase to S phase of cell cycle (https://www.ncbi.nlm.nih.gov/gene/131405). We found that the higher expression of TRIM71 was positively correlated with the higher expression of LINC002253, indicating that TRIM71 was likely to affect tumor development. Survival analysis showed that TRIM71 was highly expressed in WT patients with poor prognosis, suggesting a potential of being a tumor prognostic marker. Ren et al. pointed out that TRIM71 promoted the proliferation of non-small cell lung cancer cells through activation of the inhibitor of κB pathway (36). Another study indicated that TRIM71 could interact with p53, control its abundance by ubiquitination and decrease p53-dependent pro-apoptotic responses (37). However, the specific biological mechanism of TRIM71 in WT remained unclear and needs to be further explored.
Because mRNA functions as downstream of the network in ceRNA hypothesis, GO and KEGG analysis was conducted to excavate the biological action mechanisms and pathways of the DemRNAs. “Mitotic nuclear division” was the most enriched GO terms in BP. Its abnormity may lead to chromosome division errors which accounts for excessive cell proliferation. We speculated that it might also play a role in the occurrence of WT. Other certified tumor-related terms included “nuclear chromatin” (38), “G2/M transition of mitotic cell cycle” (39), “sister chromatid cohesion” (40), “regulation of transcription involved in G1/S transition of mitotic cell cycle” (31), “DNA repair” (41), “cell proliferation” (31) and “cell division” (42). The pathway analyses showed that various pathways were involved in cell division and proliferation, such as “DNA replication pathway”, “Oocyte meiosis pathway” and “Homologous recombination pathway”.
Admittedly, our article still has some limitations. First, due to the rarity of WT, we are currently unable to conduct the multi-center clinical verification. Second, we have not conducted in vivo and in vitro experiments so far. So, we have limited evidence to explain the mechanism of WT caused by TRIM71 in particular. Nevertheless, our study explored the most closely related genes involved in hub sub-network of WT, which could provide preliminary results for further study.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.01.34). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Saha H, Ghosh D, Biswas SK, et al. Synchronous Bilateral Wilms Tumor: Five-Year Single-Center Experience with Assessment of Quality of Life. J Indian Assoc Pediatr Surg 2019;24:52-60. [Crossref] [PubMed]
- Breslow NE, Beckwith JB, Perlman EJ, et al. Age distributions, birth weights, nephrogenic rests, and heterogeneity in the pathogenesis of Wilms tumor. Pediatr Blood Cancer 2006;47:260-7. [Crossref] [PubMed]
- Raffensperger J. Max Wilms and his tumor. J Pediatr Surg 2015;50:356-9. [Crossref] [PubMed]
- Dumoucel S, Gauthier-Villars M, Stoppa-Lyonnet D, et al. Malformations, genetic abnormalities, and Wilms tumor. Pediatr Blood Cancer 2014;61:140-4. [Crossref] [PubMed]
- Rankin J, Silf KA, Pearce MS, et al. Congenital anomaly and childhood cancer: A population-based, record linkage study. Pediatr Blood Cancer 2008;51:608-12. [Crossref] [PubMed]
- Nakayama DK, Bonasso PC. The History of Multimodal Treatment of Wilms' Tumor. Am Surg 2016;82:487-92. [PubMed]
- Green DM. The evolution of treatment for Wilms tumor. J Pediatr Surg 2013;48:14-9. [Crossref] [PubMed]
- Geller JI. Current standards of care and future directions for "high-risk" pediatric renal tumors: Anaplastic Wilms tumor and Rhabdoid tumor. Urol Oncol 2016;34:50-6. [Crossref] [PubMed]
- Mavinkurve-Groothuis AM, van den Heuvel-Eibrink MM, Tytgat GA, et al. Treatment of relapsed Wilms tumour (WT) patients: experience with topotecan. A report from the SIOP Renal Tumour Study Group (RTSG). Pediatr Blood Cancer 2015;62:598-602. [Crossref] [PubMed]
- Huang C, Yuan N, Wu L, et al. An integrated analysis for long noncoding RNAs and microRNAs with the mediated competing endogenous RNA network in papillary renal cell carcinoma. Onco Targets Ther 2017;10:4037-50. [Crossref] [PubMed]
- Huang CG, Li FX, Pan S, et al. Identification of genes associated with castrationresistant prostate cancer by gene expression profile analysis. Mol Med Rep 2017;16:6803-13. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Eissa S, Safwat M, Matboli M, et al. Measurement of Urinary Level of a Specific Competing endogenous RNA network (FOS and RCAN mRNA/ miR-324-5p, miR-4738-3p, /lncRNA miR-497-HG) Enables Diagnosis of Bladder Cancer. Urol Oncol 2019;37:292.e19-292.e27. [Crossref] [PubMed]
- Abdollahzadeh R, Daraei A, Mansoori Y, et al. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol 2019;234:10080-100. [Crossref] [PubMed]
- Shi L, Hong X, Ba L, et al. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis 2019;10:150. [Crossref] [PubMed]
- Liu J, Li H, Zheng B, et al. Competitive Endogenous RNA (ceRNA) Regulation Network of lncRNA-miRNA-mRNA in Colorectal Carcinogenesis. Dig Dis Sci 2019;64:1868-77. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011;696:291-303. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Tian F, Yourek G, Shi X, et al. The development of Wilms tumor: from WT1 and microRNA to animal models. Biochim Biophys Acta 2014;1846:180-7. [PubMed]
- Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol 2017;1008:1-46. [Crossref] [PubMed]
- Akhade VS, Pal D, Kanduri C. Long Noncoding RNA: Genome Organization and Mechanism of Action. Adv Exp Med Biol 2017;1008:47-74. [Crossref] [PubMed]
- Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci 2013;14:4655-69. [Crossref] [PubMed]
- Zhang EB, Han L, Yin DD, et al. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med Oncol 2014;31:914. [Crossref] [PubMed]
- Cui J, Mo J, Luo M, et al. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:12400-9. [PubMed]
- Li Y, Wang Z, Shi H, et al. HBXIP and LSD1 Scaffolded by lncRNA Hotair Mediate Transcriptional Activation by c-Myc. Cancer Res 2016;76:293-304. [Crossref] [PubMed]
- Sun K, Jia Z, Duan R, et al. Long non-coding RNA XIST regulates miR-106b-5p/P21 axis to suppress tumor progression in renal cell carcinoma. Biochem Biophys Res Commun 2019;510:416-20. [Crossref] [PubMed]
- Gao S, Lin Z, Li C, et al. lncINS-IGF2 Promotes Cell Proliferation and Migration by Promoting G1/S Transition in Lung Cancer. Technol Cancer Res Treat 2019;18:1533033818823029. [Crossref] [PubMed]
- Dong P, Xiong Y, Yue J, et al. Exploring lncRNA-Mediated Regulatory Networks in Endometrial Cancer Cells and the Tumor Microenvironment: Advances and Challenges. Cancers (Basel) 2019. [Crossref] [PubMed]
- Nava-Rodríguez MP, Dominguez-Cruz MD, Aguilar-Lopez LB, et al. Genomic instability in a chronic lymphocytic leukemia patient with mono-allelic deletion of the DLEU and RB1 genes. Mol Cytogenet 2019;12:2. [Crossref] [PubMed]
- Wang L, Liu D, Wu X, et al. Long non-coding RNA (LncRNA) RMST in triple-negative breast cancer (TNBC): Expression analysis and biological roles research. J Cell Physiol 2018;233:6603-12. [Crossref] [PubMed]
- Hatakeyama S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem Sci 2017;42:297-311. [Crossref] [PubMed]
- Ren H, Xu Y, Wang Q, et al. E3 ubiquitin ligase tripartite motif-containing 71 promotes the proliferation of non-small cell lung cancer through the inhibitor of kappaB-alpha/nuclear factor kappaB pathway. Oncotarget 2017;9:10880-90. [Crossref] [PubMed]
- Nguyen DTT, Richter D, Michel G, et al. The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death Differ 2017;24:1063-78. [Crossref] [PubMed]
- Hanna MG, Liu C, Rohde GK, et al. Predictive Nuclear Chromatin Characteristics of Melanoma and Dysplastic Nevi. J Pathol Inform 2017;8:15. [Crossref] [PubMed]
- Priami C, De Michele G, Cotelli F, et al. Modelling the p53/p66Shc Aging Pathway in the Shortest Living Vertebrate Nothobranchius Furzeri. Aging Dis 2015;6:95-108. [Crossref] [PubMed]
- Xie XW, Wang XY, Liao WJ, et al. Effect of Upregulated DNA Replication and Sister Chromatid Cohesion 1 Expression on Proliferation and Prognosis in Hepatocellular Carcinoma. Chin Med J (Engl) 2018;131:2827-35. [PubMed]
- Leongamornlert DA, Saunders EJ, Wakerell S, et al. Germline DNA Repair Gene Mutations in Young-onset Prostate Cancer Cases in the UK: Evidence for a More Extensive Genetic Panel. Eur Urol 2019;76:329-37. [Crossref] [PubMed]
- Tominaga K, Minato H, Murayama T, et al. Semaphorin signaling via MICAL3 induces symmetric cell division to expand breast cancer stem-like cells. Proc Natl Acad Sci U S A 2019;116:625-30. [Crossref] [PubMed]


