
Telomerase positive CTCs with PSMA high expression associated with prostate cancer metastasis
Introduction
Prostate cancer (PCa) is one of the most common cancer in men, and the incidence rate was increased in China in recent years. Radical prostatectomy (RP), hormone therapy, and radiotherapy (RT) are the treatment options for PCa (1). Although the patients with localized PCa will meet a long-time prognosis after the surgery and regular treatment, some PCa patients will progress to metastatic PCa and further develop castration-resistant PCa, which was associated with high lethality and poor prognosis (2,3). Thus, exploring the metastasis-related biomarker is critical for changing the treatment strategy to prevent metastasis events.
Circulating tumor cells (CTCs) are cancer cells detached from the primary tumor site into the circulation and regarded as potential metastatic seeds. CTCs have been reported to detect tumor metastasis and prognosis among those patients, which could be an ideal biomarker for PCa metastasis management (4-6). We have previously established a telomerase reverse transcriptase (TERT)-based CTC detection (TBCD) method and showed a high sensitivity in lung cancer diagnosis (7,8). High telomerase activity plays a crucial role in the high division activity of cancer cells, and this approach could enrich and separate viable tumor cells with high proliferation ability (9,10). Thus, further characterizing telomerase-positive CTCs may disclose the characters of the metastatic process.
Prostate-specific membrane antigen (PSMA) is a kind of glutamate carboxypeptidase expressed on the membrane surface. Some studies have shown that PSMA is expressed in most primary or metastatic PCa and positively correlated with the malignancy of cancer (11-13). Due to the high expression of PSMA in PCa and its high affinity with antibodies, some studies indicated that PSMA could play an essential role in diagnosing and treating PCa (14-16). To note, PSMA has been proved that widely correlated with PCa metastasis in recent years, and Cho et al. showed that PSMA expression was significantly higher in cases of metastatic prostate (11,17). Thus, whether the PSMA expression level on CTCs was related to the metastatic progress in PCa still needs further elucidation. In this study, bioinformatics analysis was performed to confirm that high PSMA expression CTCs were associated with tumor metastasis. Then, we further revealed the relationship between ‘TERT+ PSMA+ CTCs’ and PCa metastasis. We present the following article in accordance with the STROBE reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-21-1140/rc).
Methods
Patients and study design
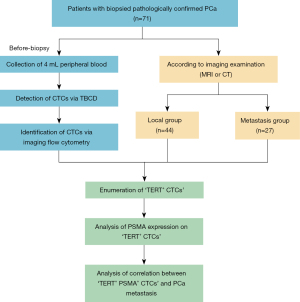
A total of 71 patients diagnosed as PCa in the Cancer Hospital, Chinese Academy of Medical Sciences were enrolled from July 2018 to May 2021. PCa was defined according to the National Comprehensive Cancer Network (NCCN) guidelines. All biopsied specimens were evaluated and diagnosed by experienced pathologists according to International Association of Uropathology (ISUP) 2014 modified Gleason score system. Metastatic PCa was defined as tumor metastasis to bone, lung, liver, or other organs according to imaging examination. Sixty-one patients had magnetic resonance imaging (MRI)-detected abnormalities. The other 10 patients only received computerized tomography (CT) due to the contraindications. MRI results were scored based on the prostate imaging and reporting and data system (PIRADS) version 2. See Figure 1 for study flowchart. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval was obtained from the ethics committees of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. NCC2014G-16). All enrolled patients have signed written informed consent.
CTCs detection reagents
CTCs were detected using a reagent of herpes simplex virus type 1 (oHSV1)-human telomerase reverse transcriptase promoter (hTERTp)-green fluorescent protein (GFP).
The oHSV1-hTERTp-GFP was designed by replacing the endogenous ICP4 promoter with a TERT promoter and replacing the genes encoding cell protein 34.5 (ICP34.5) with a GFP gene as described before (7,8). The purified viruses were stored at −80 ℃ until use.
Peripheral blood (PB) sampling and CTC identification
Collected 4 milliliters of prebiopsy PB with ethylene diamine tetraacetic acid (EDTA) tubes and kept them at 4 ℃ until separation. Sample preparation and detailed workflow were carried out as previously described (7,8). In brief, the oHSV1-hTERTp-GFP was used to infect the cells after lysis of red blood cells, then infected cells were incubated at 5% CO2 and 37 ℃ cell incubator for 24 h. The transduced cells were collected, then stained with an anti-CD45 antibody (Biolegend, San Diego, CA, USA; Cat: 304014) and anti-PSMA antibody (Biolegend, Cat: 342508). CTCs were detected by flow cytometry. The ‘CD45− GFP+ cells’ were recorded as ‘TERT+ CTCs’, the ‘CD45− GFP+ PSMA+ cells’ were recorded as ‘TERT+ PSMA+ CTCs’.
Identification of CTCs using ImageStreamX®
One tube of 4 mL PB was prepared, as described above. The sample was treated with a standard CTC identification process. The cells were collected and incubated with an eFluor 405-labeled anti-CD45 antibody (eBioscience, Waltham, MA, USA; Cat: 48-0459-42) and APC-labeled anti-PSMA antibody (Biolegend, Cat: 342508). The incubation conditions were 30 min at room temperature. After washing, CTCs were detected with an ImageStreamX® Mark II system (Amnis, Seattle, WA, USA) for imaging flow cytometry.
Bioinformatics analysis
We downloaded the data of PSMA expression profile in PCa primary tumor site, CTCs or white blood cell (WBC) from the ctcRbase database (http://www.origin-gene.cn/database/ctcRbase/index.html). The PCa CTCs single-cell expression profile data were downloaded from the ctcRbase database (http://www.origin-gene.cn/database/ctcRbase/index.html) and GEO database (GSE67980) (18).
Gene ontology (GO) functional classification and enrichment analysis were performed to identify significantly enriched GO terms for differentially expressed proteins between PSMA high expression group and the low expression group. We further extracted 27 metastasis-related gene panels (Table S1) from the literature review based on the following reasons (19-22): (I) these genes have been proven to be associated with tumor metastasis by many studies, (II) this gene panel is involved in tumor metastasis-associated signaling activation, genomic instability, cell migration and colonization, and so on. The gene expression levels were defined as the sum of the total gene expression value. Expression reads of gene panels were first normalized to fragments per kilobase of exon model per million mapped fragments (FPKM) values, then calculated and compared the sum of the expression values between PSMA high expression group and low expression group.
Statistical analysis
Data analysis in this study was performed with standard software (R, Prism GraphPad version 8.2). The nonparametric Mann-Whitney U test was used to test asymmetric and continuous data between patient groups. The expected and observed frequencies in categorical variables between two groups were analyzed using Pearson’s chi-squared test. Data are shown as mean values with standard deviations (mean ± SD). Results were considered statistically significant when the P value was less than 0.05.
Results
Patient characteristics
The clinical characteristics of the enrolled patients are provided in Table 1. A total of 71 PCa patients were enrolled in this study. According to the progress of the PCa, 44 patients were divided into local PCa groups, and 27 patients were divided into metastatic PCa groups. The median prostate-specific antigen (PSA) value in the local PCa group was 12.27 ng/mL, whereas the metastasis group had a higher median PSA value of 321.1 ng/mL (P<0.0001). Besides, metastasis PCa displayed a higher pathologic Gleason score (P=0.0092) and MRI PIRADS score (P<0.0001). In the local PCa group, 36 patients accepted the RP and 8 patients accepted the RT or RT combined with androgen deprivation therapy (ADT); in the metastasis PCa group, 19 patients accepted the ADT combined with novel hormonal agents (NHAs) and 8 patients accepted the ADT combined with chemotherapy. The pathology results of post-RP in the local PCa group were provided in Table 1. Age and family history were not statistically different between the two groups (P>0.05).
Table 1
| Characteristic | Local PCa (n=44) | Metastatic PCa (n=27) | P |
|---|---|---|---|
| Age, year (range) | 68.73 (56–82) | 69.74 (55–83) | 0.635 |
| Median PSA (range), ng/mL | 12.27 (4.48–99.12) | 321.1 (4.67–9,995) | <0.0001 |
| Gleason score, n (%) | 0.0092 | ||
| 3+3 | 8 (18.2) | – | |
| 3+4 | 5 (11.4) | – | |
| 4+3 | 6 (13.6) | 2 (7.4) | |
| 4+4 | 18 (40.9) | 14 (51.9) | |
| 4+5 | 5 (11.4) | 5 (18.5) | |
| 5+4 | 1 (2.3) | 6 (22.2) | |
| 5+5 | 1 (2.3) | – | |
| Imaging test, n (%) | – | ||
| MRI | 44 (100.0) | 17 (63.0) | |
| CT | 0 | 10 (37.0) | |
| MRI score, n (%) | <0.0001 | ||
| 3 | 18 (40.9) | – | |
| 4 | 16 (36.4) | 1 (5.9) | |
| 5 | 10 (22.7) | 16 (94.1) | |
| Treatment, n (%) | – | ||
| RP | 36 (81.8) | – | |
| RT or RT + ADT | 8 (18.2) | – | |
| ADT + NHAs | – | 19 (70.4) | |
| ADT + chemotherapy | – | 8 (29.6) | |
| pT, n (%) | – | ||
| pT2 | 24 (66.7) | – | |
| pT3 | 12 (33.3) | – | |
| pT4 | 0 | – | |
| pN, n (%) | – | ||
| pN0 | 1 (2.8) | – | |
| pN1 | 35 (97.2) | – | |
| Family history | None | None | – |
PCa, prostate cancer; PSA, prostate-specific antigen; MRI, magnetic resonance imaging; CT, computerized tomography; RP, radical prostatectomy; RT, radiotherapy; ADT, androgen deprivation therapy; NHAs, novel hormonal agents; pT, pathological tumor; pN, pathological node.
Correlation between telomerase positive CTCs and clinical characteristics
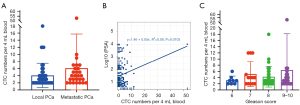
First, we analyzed the correlation between TERT-positive CTCs and clinical features. As shown in Figure 2A, the mean CTC number was 6.11±9.63 per 4 mL blood in the metastasis PCa group and 4.09±3.41 per 4 mL blood in the local PCa group, there was no statistical difference between the two groups (P=0.11). There was also no significant linear correlation between PSA level and CTC numbers (Figure 2B). Furthermore, PCa with higher grades of Gleason score (score 6 vs. score 7 vs. score 8 vs. score 9–10 = 3.13±1.36 vs. 5.15±4.04 vs. 4.28±3.34 vs. 6.44±11.74, P=0.79) tend to detect more CTCs (Figure 2C), although there was no statistical difference.
PSMA was highly expressed in CTCs and associated with PCa metastasis
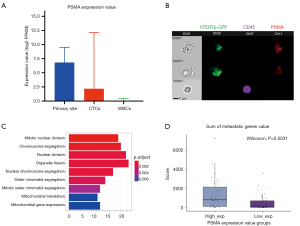
To uncover the correlation between CTCs and PSMA expression, we first analyzed 77 CTCs’ single-cells sequencing data. The results showed that the PSMA was a higher expression in the PCa primary site (median =6.81, range from 0.64 to 9.53) and CTC (median =2.18, range from 0 to 12.12) than the WBC (median =0, range from 0 to 0.49) (P<0.05) (Figure 3A). Subsequently, we applied Flowsight to confirm the PSMA was expressed in CTCs. As shown in Figure 3B, CTCs highly expressed GFP and PSMA, and without CD45 expression. WBCs expressed only CD45. On the other hand, CTCs were also larger than WBCs. To analyze whether the PSMA high expression rate can promote the metastatic capability of PCa CTC, we divided 77 PCa CTCs single-cell expression profile data into PSMA high expression group (PSMA-high group) and PSMA low expression group (PSMA-low group) based on the median value of PSMA expression. We performed GO enrichment analysis of differentially expressed genes between two groups. As shown in Figure 3C, GO analysis revealed that the major terms of cell division related to proliferation were enriched in the PSMA-high group. To further demonstrate the relationship between proliferation and metastasis, we used a 27 metastasis-related genes panel (Table S1) to verify it. As shown in Figure 3D, the sum of genes expression value was significantly higher in the PSMA-high group than the PSMA-low group (P=0.0031). These results suggest that high expression of PSMA in CTCs may be associated with the metastasis of PCa.
PSMA and TERT double-positive CTCs was closely related to the metastasis of PCa
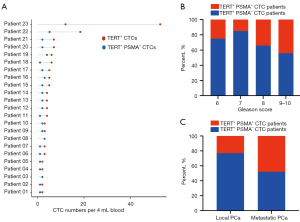
We further used TBCD to confirm that PSMA high-expressed CTCs were correlated with the metastatic events. As shown in Figure 4A, 23 out of 71 patients detected at least one PSMA positive CTCs, and we defined those patients as ‘TERT+ PSMA+ CTCs’ patients, and other patients as ‘TERT+ PSMA− CTCs’ patients. We first depicted the distribution of ‘TERT+ PSMA+ CTCs’ patients among the different Gleason scores representing PCa risk classification. In high Gleason score patients (Gleason >7), ‘TERT+ PSMA+ CTCs’ patient’s positive rate was significantly higher than in intermediate and low Gleason score patients (Gleason score ≤7) (score 6 vs. score 7 vs. score 8 vs. score 9–10 = 25.00% vs. 15.28% vs. 34.38% vs. 44.44%, P<0.0001, Figure 4B). The data indicated that more ‘TERT+ CTC’ with PSMA high expression could be detected in higher-risk PCa. Finally, we compared the proportion of ‘TERT+ PSMA+ CTCs’ patients between the local PCa group and metastatic PCa group. As shown in Figure 4C, the percent of ‘TERT+ PSMA+ CTCs’ patients was 48.15% (13 out of 27 patients) in the metastatic group, significantly higher than 22.72% (10 out of 44 patients) in the local group (P=0.0263). These data suggested that TERT and PSMA double-positive CTCs are closely related to the metastasis of PCa.
Discussion
In this study, we used TBCD to detect the expression rate of PSMA on telomerase-positive CTCs between the local PCa group and metastatic PCa group, and provided new evidence that PSMA and TERT double-positive CTCs were associated with the high tumor risk and metastasis.
CTCs were shedding from the primary tumor into the PB circulation and potentially functioning as seeds to form the metastatic foci (4,23). Food and Drug Administration (FDA) has approved CTC-enumeration as an indicator for the evaluation of PCa metastasis burden. Malihi et al. (24) proved that CTCs with genomic instability could be the aggressive hallmark of advanced PCa. Some studies revealed that the CTCs with diverse phenotypes are closely related to cancer progression and metastasis (25,26). It is of great importance for prognosis evaluation through CTCs detection among advanced PCa patients, and we also confirmed that CTCs could be an ideal marker to predict tumor metastasis.
CTCs as a promising tool to develop predictive molecular biomarkers require more advanced and sophisticated equipment. Currently, CTCs enrichment is based on technologies as below: (I) CTC-specific ligand-dependent method: microfluidic system, CellSearch, immunomagnetic enrichment, etc. (II) CTC-specific ligand-independent (physics-based) methods: size, deformability-, density-based detection, etc. (27). EpCAM was a promising epithelial cell-specific marker for isolating CTCs. However, CTCs may lose EpCAM-specific molecules due to the epithelial-mesenchymal transition (EMT) process, resulting in the inability to enrich CTCs with mesenchymal phenotypes (28). Those CTCs underwent the EMT may represent a more malignant subset of tumor cells. Some tumor cells with weak or no expression of EpCAM antigens also cannot be effectively used (29). On the other hand, some benign epithelial circulating cells may contaminate CTCs and cause false positives. Those weak points may limit EpCAM’s clinical application. Telomerase is a pan tumor hallmark which highly expressed in about 80% of malignant tumors and is required for malignant transformation and tumor progression (9). Some studies indicated that tumors with higher telomerase activity are associated with activation of energy metabolism, stem cell activity and cell migration (10,30). Accordingly, telomerase-based CTC detection is a promising approach to capture living CTCs for tumor progression monitoring, tumor diagnosing and therapeutic efficacy evaluation (10). Hwang et al., Ma et al., Togo et al. and Takakura et al. (31-34) conducted various studies to confirm that telomerase-based CTC detection is a powerful weapon for CTC detection in PCa, breast cancer, pancreatic cancer, non-small cell lung cancer, and cervical cancer. We also applied TBCD in diagnosing early PCa and pulmonary nodules and exhibited a high diagnostic ability. Besides, a very low amount ‘GFP+ CD45−’ background cells was detected for benign prostate disease (1.15±1.87 cells per 4 mL PB), benign lung nodules (1.29±0.43 cells per 4 mL PB), or healthy donors (0.63 per 4 mL PB) (7,8,35). Fewer studies further exploited its clinical application for tumor metastasis prediction. In this study, we used TBCD to detect the viable CTCs between the local PCa group and metastatic PCa group, and showed that the mean number of TERT-based CTCs in the metastatic group was higher than the local group. However, there is no statistical difference between the two groups, which may due to the limitation of sample size.
As a high expression pattern antigen expressed on the PCa membrane surface, PSMA-specific probe positron emission computed tomography (PET) was clinically widely used for cancer lesion visualization and confirmed the value in prostate disease diagnosing, staging and treatment in some studies (11,14-16). Furthermore, PSMA was reported to promote cancer progression and metastatic lesion formation (11). Our study provided reliable evidence that increased PSMA expression in PCa primary lesions and CTCs compared to normal prostate tissue via ctcRdatabase, and confirmed that CTCs with high PSMA expression had stronger proliferation and metastasis ability. PSMA expression was reported to correlate with Gleason score and cancer aggressiveness, which is also shown in our results (36). Besides, some studies showed that PSMA high expression CTCs were associated with short overall survival and progression free survival (PFS) in metastatic PCa (37-39). However, those studies above did not provide evidence that PSMA was associated with tumor metastasis. In our study, we showed that ‘TERT+ PSMA+ CTCs’ was associated with PCa metastasis which implies that those CTCs could be the ‘seeds’ of metastatic site. We believe that ‘TERT+ PSMA+ CTCs’ could be the concomitant diagnosis for PSMA-PET and drug therapy to predict metastasis. These patients with PSMA+ CTCs should be closely followed up in clinical practice. Therefore, we will also follow up those patients with PSMA-positive CTCs in the local group to further extend the significance of our study.
Conclusions
Our study suggested that telomerase positive CTCs with high PSMA expression were associated with the PCa metastatic progress.
Acknowledgments
We thank the staff of the National Center for Protein Sciences Beijing (Peking University) and Ms. Fei Wang for her technical assistance with the flow cytometry imaging. We also thank Diatech Co. for the technical support for the TBCD method.
Funding: This work was supported by the National Key Research and Development Project (No. 2019YFC1315700 and 2017YFC1308702) and the CAMS Initiative for Innovative Medicine (No. 2017-I2M-1-005).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-21-1140/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-21-1140/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-21-1140/coif). CZ is an employee of Chongqing Diatech Biotechnological Limited Company. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval was obtained from the ethics committees of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. NCC2014G-16). All enrolled patients have signed written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Litwin MS, Tan HJ. The Diagnosis and Treatment of Prostate Cancer: A Review. JAMA 2017;317:2532-42. [Crossref] [PubMed]
- Teo MY, Rathkopf DE, Kantoff P. Treatment of Advanced Prostate Cancer. Annu Rev Med 2019;70:479-99. [Crossref] [PubMed]
- Komura K, Sweeney CJ, Inamoto T, et al. Current treatment strategies for advanced prostate cancer. Int J Urol 2018;25:220-31. [Crossref] [PubMed]
- Castro-Giner F, Aceto N. Tracking cancer progression: from circulating tumor cells to metastasis. Genome Med 2020;12:31. [Crossref] [PubMed]
- Xu L, Mao X, Grey A, et al. Noninvasive Detection of Clinically Significant Prostate Cancer Using Circulating Tumor Cells. J Urol 2020;203:73-82. [Crossref] [PubMed]
- Pantel K, Hille C, Scher HI. Circulating Tumor Cells in Prostate Cancer: From Discovery to Clinical Utility. Clin Chem 2019;65:87-99. [Crossref] [PubMed]
- Zhang W, Duan X, Zhang Z, et al. Combination of CT and telomerase+ circulating tumor cells improves diagnosis of small pulmonary nodules. JCI Insight 2021;6:e148182. [Crossref] [PubMed]
- Zhang W, Bao L, Yang S, et al. Tumor-selective replication herpes simplex virus-based technology significantly improves clinical detection and prognostication of viable circulating tumor cells. Oncotarget 2016;7:39768-83. [Crossref] [PubMed]
- Dratwa M, Wysoczańska B, Łacina P, et al. TERT-Regulation and Roles in Cancer Formation. Front Immunol 2020;11:589929. [Crossref] [PubMed]
- Soria E, Vallejo M, Saenz L, et al. Telomerase-specific attenuated viruses, a definitive strategy or just one more in circulating tumor cells detection anthology? Cancer Lett 2020;469:490-7. [Crossref] [PubMed]
- Lütje S, Heskamp S, Cornelissen AS, et al. PSMA Ligands for Radionuclide Imaging and Therapy of Prostate Cancer: Clinical Status. Theranostics 2015;5:1388-401. [Crossref] [PubMed]
- Ananias HJ, van den Heuvel MC, Helfrich W, et al. Expression of the gastrin-releasing peptide receptor, the prostate stem cell antigen and the prostate-specific membrane antigen in lymph node and bone metastases of prostate cancer. Prostate 2009;69:1101-8. [Crossref] [PubMed]
- Paschalis A, Sheehan B, Riisnaes R, et al. Prostate-specific Membrane Antigen Heterogeneity and DNA Repair Defects in Prostate Cancer. Eur Urol 2019;76:469-78. [Crossref] [PubMed]
- Eiber M, Fendler WP, Rowe SP, et al. Prostate-Specific Membrane Antigen Ligands for Imaging and Therapy. J Nucl Med 2017;58:67S-76S. [Crossref] [PubMed]
- Petrylak DP, Kantoff P, Vogelzang NJ, et al. Phase 1 study of PSMA ADC, an antibody-drug conjugate targeting prostate-specific membrane antigen, in chemotherapy-refractory prostate cancer. Prostate 2019;79:604-13. [Crossref] [PubMed]
- Powers E, Karachaliou GS, Kao C, et al. Novel therapies are changing treatment paradigms in metastatic prostate cancer. J Hematol Oncol 2020;13:144. [Crossref] [PubMed]
- Cho H, Chung JI, Kim J, et al. Multigene model for predicting metastatic prostate cancer using circulating tumor cells by microfluidic magnetophoresis. Cancer Sci 2021;112:859-70. [Crossref] [PubMed]
- Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015;349:1351-6. [Crossref] [PubMed]
- Wang G, Zhao D, Spring DJ, et al. Genetics and biology of prostate cancer. Genes Dev 2018;32:1105-40. [Crossref] [PubMed]
- Grimm D, Bauer J, Wise P, et al. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol 2020;67:122-53. [Crossref] [PubMed]
- Das SK, Maji S, Wechman SL, et al. MDA-9/Syntenin (SDCBP): Novel gene and therapeutic target for cancer metastasis. Pharmacol Res 2020;155:104695. [Crossref] [PubMed]
- Sun Y, Ma L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers (Basel) 2019;11:216. [Crossref] [PubMed]
- Gkountela S, Castro-Giner F, Szczerba BM, et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019;176:98-112.e14. [Crossref] [PubMed]
- Malihi PD, Graf RP, Rodriguez A, et al. Single-Cell Circulating Tumor Cell Analysis Reveals Genomic Instability as a Distinctive Feature of Aggressive Prostate Cancer. Clin Cancer Res 2020;26:4143-53. [Crossref] [PubMed]
- Lindsay CR, Le Moulec S, Billiot F, et al. Vimentin and Ki67 expression in circulating tumour cells derived from castrate-resistant prostate cancer. BMC Cancer 2016;16:168. [Crossref] [PubMed]
- Bastos DA, Antonarakis ES. CTC-derived AR-V7 detection as a prognostic and predictive biomarker in advanced prostate cancer. Expert Rev Mol Diagn 2018;18:155-63. [Crossref] [PubMed]
- Shen Z, Wu A, Chen X. Current detection technologies for circulating tumor cells. Chem Soc Rev 2017;46:2038-56. [Crossref] [PubMed]
- Gorges TM, Tinhofer I, Drosch M, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012;12:178. [Crossref] [PubMed]
- Diéguez L, Winter MA, Pocock KJ, et al. Efficient microfluidic negative enrichment of circulating tumor cells in blood using roughened PDMS. Analyst 2015;140:3565-72. [Crossref] [PubMed]
- Lamb R, Ozsvari B, Bonuccelli G, et al. Dissecting tumor metabolic heterogeneity: Telomerase and large cell size metabolically define a sub-population of stem-like, mitochondrial-rich, cancer cells. Oncotarget 2015;6:21892-905. [Crossref] [PubMed]
- Hwang JE, Joung JY, Shin SP, et al. Ad5/35E1aPSESE4: A novel approach to marking circulating prostate tumor cells with a replication competent adenovirus controlled by PSA/PSMA transcription regulatory elements. Cancer Lett 2016;372:57-64. [Crossref] [PubMed]
- Ma Y, Hao S, Wang S, et al. A Combinatory Strategy for Detection of Live CTCs Using Microfiltration and a New Telomerase-Selective Adenovirus. Mol Cancer Ther 2015;14:835-43. Erratum in: Mol Cancer Ther 2015;14:1761. [Crossref] [PubMed]
- Togo S, Katagiri N, Namba Y, et al. Sensitive detection of viable circulating tumor cells using a novel conditionally telomerase-selective replicating adenovirus in non-small cell lung cancer patients. Oncotarget 2017;8:34884-95. [Crossref] [PubMed]
- Takakura M, Matsumoto T, Nakamura M, et al. Detection of circulating tumor cells in cervical cancer using a conditionally replicative adenovirus targeting telomerase-positive cells. Cancer Sci 2018;109:231-40. [Crossref] [PubMed]
- Yang Z, Bai H, Hu L, et al. Improving the diagnosis of prostate cancer by telomerase-positive circulating tumor cells: A prospective pilot study. EClinicalMedicine 2022;43:101161. [Crossref] [PubMed]
- Kratochwil C, Afshar-Oromieh A, Kopka K, et al. Current Status of Prostate-Specific Membrane Antigen Targeting in Nuclear Medicine: Clinical Translation of Chelator Containing Prostate-Specific Membrane Antigen Ligands Into Diagnostics and Therapy for Prostate Cancer. Semin Nucl Med 2016;46:405-18. [Crossref] [PubMed]
- Nagaya N, Nagata M, Lu Y, et al. Prostate-specific membrane antigen in circulating tumor cells is a new poor prognostic marker for castration-resistant prostate cancer. PLoS One 2020;15:e0226219. [Crossref] [PubMed]
- Autio KA, Dreicer R, Anderson J, et al. Safety and Efficacy of BIND-014, a Docetaxel Nanoparticle Targeting Prostate-Specific Membrane Antigen for Patients With Metastatic Castration-Resistant Prostate Cancer: A Phase 2 Clinical Trial. JAMA Oncol 2018;4:1344-51. [Crossref] [PubMed]
- Francolini G, Loi M, Salvestrini V, et al. Prospective assessment of AR splice variant and PSMA detection on circulating tumor cells of mCRPC patients: preliminary analysis of patients enrolled in PRIMERA trial (NCT04188275). Clin Exp Metastasis 2021;38:451-8. [Crossref] [PubMed]


