
Bioinformatics analysis based on immune-autophagy-related lncRNAs combined with immune infiltration in bladder cancer
Introduction
Bladder cancer is one of the most common malignant tumors of the urinary system, with 430 thousand new cases every year (1). About 25% of the patients with myometrial invasive bladder cancer have a poor prognosis and lack effective treatment (2-4). With the progression of cancer, the 5-years survival rate of patients decreases. In patients with advanced-stage, the mortality rate is more than 50% (5). Therefore, the early evaluation of the prognosis of patients with bladder cancer has important clinical significance.
Clinical features, TMN stage, and lymph node status are critical predictive factors for bladder cancer (6,7). Previous studies have shown that bladder cancer patients with poor clinical conditions, high stage grade, and positive lymph node metastasis have lower OS (8,9). However, traditional predictive methods are mainly based on clinical and pathological information while not considering biological heterogeneity. Recently, new prognostic models have been developed based on glycolysis-related mRNAs, immune-related mRNAs, extracellular matrix-related long noncoding RNA (lncRNAs), microenvironment-related lncRNAs, tumor-infiltrating immune cells, alternative splicing events (10-17).
LncRNA is a non-protein-coding transcript consisting of more than 200 nucleotides (18,19). LncRNAs regulate the occurrence and development of tumors by binding to nuclear acids, proteins, and other macromolecules (20-24). As a result, lncRNAs can also predict cancer patients' outcomes (25,26). Although some molecular biomarkers have been used as independent prognostic factors by traditional predictive models of bladder cancer, there are some limitations, such as small sample size, different detection platforms, single prognostic variates, and similar (6,27). Therefore, it is an urgent clinical task to determine reliable prognostic biomarkers.
The TCGA database works on applying high-throughput genome analysis techniques to better understand the occurrence and progression of carcinoma, thereby improving its diagnosis, treatment, and prevention. Autophagy is a regulatory mechanism that removes unnecessary or dysfunctional cellular components and circulates metabolic substrates. In response to pressure signals in the tumor microenvironment (TME), autophagy pathways in tumor cells and immune cells are altered, thus affecting tumor progression, immunity, and treatment differently. Autophagy is an important mechanism in cancer biology, immunology, and therapy. Thus, in this study, we used a rigorous computational framework to mine immune-autophagy-related long noncoding RNA (IArlncRNAs) and clinical data of bladder cancer downloaded from the TCGA database, as well as to construct a prognostic model IArlncRNAs in order to predict the OS of bladder cancer patients and identify the potential mechanisms. We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/tau-21-560).
Methods
Download and organize transcriptional group data and clinical data
The TCGA database is a project supervised by the National Cancer Institute and National Human Genome Research Institute. In this study, we used a TCGA database (https://portal.gdc.cancer.gov/) to download transcriptome (FPKM) and clinical data of patients with bladder cancer (containing primary and metastatic bladder cancer). We organized the transcriptome data and converted the ENSG number to symbol ID. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of IArlncRNAs associated with prognosis of bladder cancer patients
The transcriptome data were divided into mRNAs and lncRNAs. IMMPORT database (https://www.immport.org) was searched for immune-related genes, as well as Human Autophagy Database (http://www.autophagy.lu/) for autophagy genes. We then identified the expression of immune-related and autophagy-related genes from the mRNA dataset by taking intersection with the two target gene sets. We also identified the immune-related lncRNAs and autophagy-related lncRNAs by the co-expression networks by using the R package “limma (version 3.46.0)” (|correlation coefficient| >0.4, and P<0.001) (28). To identify the differential expression of immune-related lncRNAs (DEir-lncRNAs) and autophagy-related (DEar-lncRNAs) lncRNAs, we performed the differential expression analysis by R package limma. Then, IArlncRNAs were identified by taking an intersection between the DEir-lncRNAs and DEar-lncRNAs.
Construction of the prognostic model
Patients who were followed-up for less than 30 days from the clinical data set were excluded. Then, we combined the expression of IArlncRNAs and survival time utilizing the intersection. According to 1:1 ratio, the total samples were randomly divided into the training group and the test group. IArlncRNAs associated with survival were screened in the training group by univariate Cox regression and LASSO regression analysis. The prognostic model was constructed based on the IArlncRNAs above by multivariate Cox regression model, and their coefficients were calculated respectively (we used the R packages “survival” and “survminer”). Then, we calculated the risk score based on the risk coefficient and expression of IArlncRNAs. The formula for calculating the risk score is listed below: Risk score = βgene 1 × exprgene 1 + βgene 2 × exprgene 2 +...+ βgene n × exprgene n.
The patients were divided into the high- and low-risk groups according to the median risk score, after which the heatmaps of IArlncRNAs associated with prognosis in the training group and test group were drawn, respectively. To identify the survival differences between the high- and low-risk groups, we drew the Kaplan-Meier survival curves, survival status distributed plots, and risk score distributed plots. Finally, we verified the predictive value of the risk score model by drawing the ROC curve of the training group and test group (we used the R package “timeROC”).
Establishment of IArlncRNA-mRNA co-expression network
To investigate the association between the IArlncRNAs and their target mRNA, the IArlncRNA-mRNA network was constructed. Pearson correlation coefficients were used to analyze the difference between IArlncRNAs and mRNAs, while the absolute threshold coefficient value >0.4 and P value <0.001 were set as significant. The IArlncRNAs-mRNAs network was visualized by the Cytoscape software (version 3.7.2, http://www.cytoscape.org/).
Independent prognostic analysis and construction of nomogram
We constructed a nomogram by incorporating the related clinical factors (age, gender, stage grade) and risk score obtained from the previous multivariate Cox regression analysis (we used the R package “rms”, “foreign”, and “survival”). We drew the calibration curves and multivariate ROC curves to evaluate the model’s discrimination and accuracy (we used the R package “timeROC”).
CIBERSORT estimation and independent prognostic analysis
To evaluate the proportion of 22 immune-infiltrating cells in each sample, we used a wildly proposed computational algorithm “Cell type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT)” in R software. The samples with P value <0.05 were considered for further analysis. According to the risk score based on the risk coefficient and expression of IArlncRNAs, we divided the samples into high- and low-risk groups. Then, the Wilcoxon test was used to identify the proportion of immune cell subtypes between the high- and low-risk groups. We identified immune-infiltrating cells associated with survival by univariate Cox regression and multivariate Cox regression (we used the R packages “survival” and “survminer”). To identify the correlation between immune-infiltrating cells and IArlncRNAs, we calculated the Pearson correlation coefficients.
Prediction of the sensitivity of chemotherapeutic agents
To evaluate the differential sensitivity of chemotherapeutic agents between the high- and low-risk groups, pRRophetic algorithm was applied to predict the drugs IC50 based on the Cancer Cell Line Encyclopedia (CCLE) (29,30). R package “pRRophetic” was used to construct the ridge regression model based on the data from the CCLE for the inserted drugs. Besides, we drew boxplots of differential IC50 of interested drugs between the high- and low-risk groups with R software (Wilcoxon signed-rank test).
Statistical analysis
We used R software (version 3.46.0) to perform all statistical analyses, including R packages limma, survival, survminer, timeROC, rms, foreign, and pRRophetic. The prognostic IArlncRNAs were screened by univariate Cox regression, LASSO regression, and multivariate Cox regression analysis. The immune-infiltrating cells associated with survival by univariate Cox regression and multivariate Cox regression. The correlation was determined by Pearson correlation analysis. OS was calculated by the Kaplan-Meier method and evaluated by the log-rank test. The Wilcoxon test was used to identify the proportion of immune cell subtypes between the high- and low-risk groups. Two-tailed P<0.05 was considered statistically significant.
Results
Characteristics of data
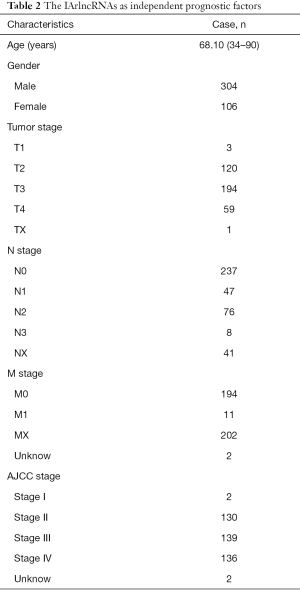
Transcriptome data and clinical data on bladder cancer were obtained from the TCGA database. There were 430 specimens in transcriptome data, including 411 tumor specimens and 19 normal specimens. The clinical data of 409 patients included 303 males, 106 females, and 158 deaths. Clinical data showed that the patients’ age ranged from 34 to 90, and their survival and follow-up time ranged from 13 to 5,050 days. Stage grade included 2 stage I, 130 stage II, 139 stage III, and 138 stage IV. These data are shown in Table 1.
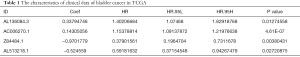
Full table
Screening of IArlncRNAs
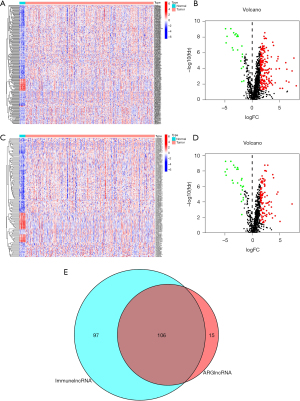
Firstly, we divided the transcriptome data downloaded from the TCGA database into mRNA dataset and lncRNA dataset. Then, we identified the immune-related genes and screened out immune-related lncRNAs by co-expression analysis (correlation coefficient >0.4, and P<0.001), as well as autophagy-related genes and autophagy-related lncRNAs (correlation coefficient >0.4, and P<0.001). Consequently, differential expression analysis of immune-related lncRNAs and autophagy-related lncRNAs between tumor and normal samples was performed (Figure 1A-D). We obtained 203 DEir-lncRNAs and 121 DEar-lncRNAs. Subsequently, Venn analysis between the DEir-lncRNAs and Dear-lncRNAs was performed, generating 106 IArlncRNAs (Figure 1E). For further analysis, patients with follow-up time <30 days were excluded. The other samples were randomly divided into two groups, including the training group (n=197) and test group (n=196). We further screened out the IArlncRNAs associated with bladder cancer patients’ prognosis in the training group by univariate Cox regression analysis and LASSO regression analysis. We determined a total of 9 IArlncRNAs by univariate Cox regression analysis. The LASSO regression analysis was performed to avoid overfitting of the model. Six IArlncRNAs were more closely related to prognosis (Figure 2A,B). Subsequently, the multivariate Cox regression analysis was performed by using the significant prognostic factors from the LASSO regression analysis. A total of 4 IArlncRNAs as independent predictors were found, including AL136084.3, AC006270.1, Z84484.1, and AL513218.1 (Table 2, Figure 2C). A correlation coefficient >0 suggested that the expression of lncRNAs were risk factors for the progression of bladder cancer, while the correlation coefficient <0 was interpreted as a protective factor.
Full table
A risk score of IArlncRNAs
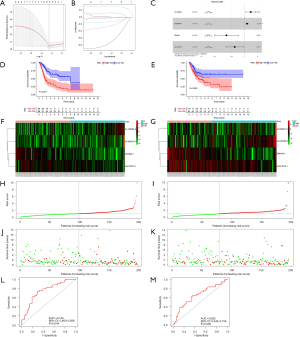
According to the correlation coefficient and expression of IArlncRNAs, we calculated the risk score for each patient by the formula. The higher the risk score, the worse the prognosis. The patients were divided into the high- and low-risk groups by the median risk score. The results showed that the OS of patients in the high-risk group was significantly worse compared to the low-risk group in the training group (Figure 2D) and in the test group (Figure 2E). The heatmap of 4 IArlncRNAs associated with bladder cancer patients’ prognosis showed differential expression of IArlncRNAs between the high- and low-risk groups in the training group (Figure 2F) and in the test group (Figure 2G). The risk score of each patient in the training and test groups are shown in Figure 2H,I. The scatterplot showed the survival status of each bladder cancer patient with a different risk score in the training group (Figure 2J) and in the test group (Figure 2K).
To evaluate the sensitivity and specificity of the risk score in predicting the survival of bladder cancer patients, we drew a time-dependent ROC curve. The AUCs of 1-year OS were 0.745 (95% CI, 0.660–0.838) in the training group (Figure 2L) and 0.652 (95% CI, 0.543–0.756) in the test group (Figure 2M).
Establishment of the lncRNA-mRNA co-expression network
Subsequently, we identified the potential functions of 4 IArlncRNAs in bladder cancer by establishing the lncRNA-mRNA co-expression network and visualizing it with Cytoscape (Figure 3A). The lncRNA-mRNA co-expression network contained 102 pairs lncRNA-mRNA with | coefficient value | >0.4 and P value <0.05. Among them, 102 mRNAs were significantly associated with 4 IArlncRNAs in the immunity and autophagy signatures. Besides, we drew a Sankey diagram to describe the risk/protective relationship between the 4 IArlncRNAs and 102 mRNAs (Figure 3B).
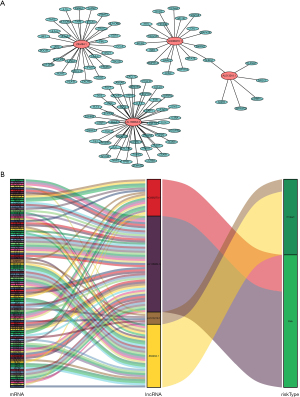
Evaluation of 4 IArlncRNAs risk score as independent factors in bladder cancer patients and the construction of the nomogram
Univariate Cox regression and multivariate Cox regression were used to identify whether the age, gender, stage, and risk score were independent prognostic factors of bladder cancer patients. The 95% CI hazard ratio of the risk score in the training group by the univariate Cox regression was 1.038 (1.020–1.056, P<0.001, Figure 4A), while the 95% CI hazard ratio of risk score by the multivariate Cox regression was 1.036 (1.018–1.054, P<0.001, Figure 4B). The 95%CI hazard ratio of the risk score in the test group by the univariate Cox regression was 1.092 (0.979–1.217, P=0.113, Figure 4C), while the 95% CI hazard ratio of risk score by the multivariate Cox regression was 1.144 (1.012–1.293, P=0.032, Figure 4D). We found that the risk score of 4 IArlncRNAs was an independent factor for bladder cancer patients.
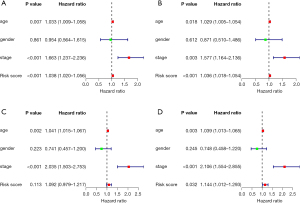
Our nomogram incorporates age, gender, stage, and risk score to predict 1-, 3-, and 5-year overall survival in patients with bladder cancer (Figure 5A). The higher the score, the worse the prognosis. Our results revealed that the risk score had the greatest impact on the prediction of overall survival. In the training group, the C index was 0.64. Also, the calibration curve and ROC curve were drawn (Figure 5B,C). The prediction was consistent with the actual observation. Similar results were found in the test group (Figure 5D-F).
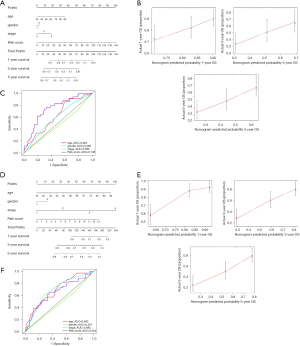
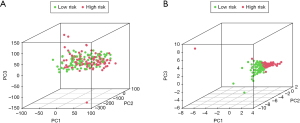
Next, we made principal components analysis (PCA) according to IArlncRNAs and total genes’ expression profiles to further understand the difference between the high- and low-risk groups. We found that the risk score can better divide the total samples into two sections, while the cumulative proportion of PC1, PC2, and PC3 were 34.39%, 61.39%, and 83.11% (Figure 6A,B).
Analysis of the composition and difference of infiltrating immune cells
We used the CIBERSORT algorithm to calculate the constituent proportion of 22 immune cell subtypes in the total samples (Figure 7A). Interestingly, B cells naïve, T cells CD8, T cells CD4 memory resting, T cells follicular helper, macrophages M1, dendritic cells resting, and dendritic cells activated had higher infiltrations in the low-risk groups, while the macrophages M2 infiltration was lower (Wilcoxon test, all P<0.05) (Figure 7B). Subsequently, the immune cell subtypes related to survival were screened by univariate Cox and multivariate Cox regression analysis, while M2 type macrophages and neutrophils were independent prognostic factors (Figure 7C).
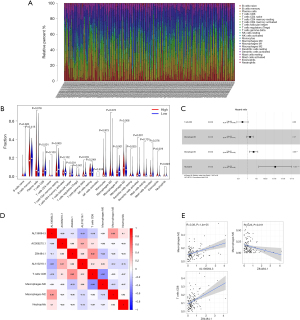
We performed co-expression analysis between the 4 prognostic IArlncRNAs and proportions of infiltrating immune cells using the Pearson correlation coefficient (Figure 7D). The fraction of Macrophages M2 was positively associated with AL136084.3 (R=0.38, P=1.4e-05). The fraction of T cells CD8 was positively associated with Z84484.1 (R=0.42, P=9.1e-07). The fraction of macrophages M0 was negatively associated with Z84484.1 (R=−0.23, P=0.011) (Figure 7E).
Differential response of chemotherapy upon high- and low-risk patients
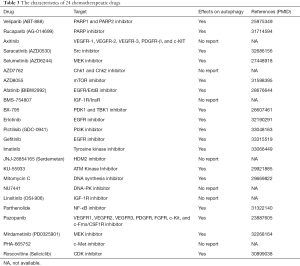
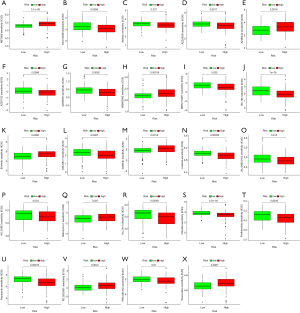
To investigate the differential IC50 of 138 chemotherapeutic drugs between the high- and low-risk groups, pRRophetic algorithm was applied to data from the CCLE. We identified 24 chemotherapeutic drugs containing a variety of anticancer mechanisms (Table 3). Besides, we described their targets, whether they have an effect on autophagy of cancer cells, and their references. Further, the boxplots for the differential IC50 of 24 chemotherapeutic drugs between the high- and low-risk groups were drawn (Figure 8A-X, P<0.05).
Full table
Discussion
Bladder cancer is a common malignant tumor of the urinary system with a high recurrence rate and different progression rates. At present, the most common treatments for bladder cancer include surgery, radiotherapy, and chemotherapy. The progressive development of new treatment strategies has significantly improved the survival rate of bladder cancer patients. However, the risk of mortality in patients with advanced bladder cancer is still high, and there is a lack of an excellent prognostic prediction model. At present, postoperative follow-up monitoring of the recurrence of bladder cancer patients mainly depends on cystoscopy. Yet, this invasive method limits its application in large-scale cancer surveillance. In addition, urine cytology is poorly used in predicting low-grade bladder cancer. Identifying biomarkers of bladder cancer may improve the ability to screen and diagnose bladder cancer and judge its prognosis. Therefore, there is an urgent requirement to explore the novel and effective prognostic indicators of bladder cancer.
Bioinformatics technology is a powerful high-throughput tool for screening molecular biomarkers and prognostic indicators. LncRNA is a non-protein-coding transcript with a length of more than 200 nucleotides and different functions in different physiological and pathological conditions. LncRNAs have tissue-specific expression patterns and a vital role in various diseases, including cancer. For example, Zhan et al. (31) found that SOX2OT is positively correlated with histological grade, TNM stage, and poor prognosis and may serve as a potential prognostic factor in bladder cancer. Moreover, Zheng et al. (32) suggested lncRNA PTENP1 as a promising biomarker for detecting bladder cancer; lncRNA PTENP1 has a suppressor role in cancer progression and promotes prognosis. Besides, Zhan et al. (33) also showed an interesting result with a great reference value that PCAT-1 acts as an independent prognostic factor for recurrence-free survival of bladder cancer and presents a potential value for clinical diagnosis and prognosis.
In the present study, we identified 4 IArlncRNAs with immunity and autophagy signatures as independent factors that contributed to the survival of bladder cancer patients, including AL136084.3, AC006270.1, Z84484.1, and AL513218.1. According to the risk score, the patients were divided into high- and low-risk groups and analyzed the two groups’ immune infiltration. We found that the proportion of B cells naïve, T cells CD8, T cells CD4 memory resting, T cells follicular helper, macrophages M1, dendritic resting and activated cells, and macrophages M2 were different between the two groups, which may contribute to the regulation of immunity. Endogenous tumor-associated antigens (TAA), recognized and presented to T lymphocytes by dendritic cells (DCs) with the major histocompatibility complex (MHC), leads to the adaptive anti-cancer immune responses (34,35). Classic MHC includes MHC I, MHC II, and MHC III; when MHC I presents self-proteins and viral proteins to CD8+ T lymphocytes, MHC II presents extracellular antigens to CD4+ T lymphocytes. In many types of tumor cells, MHC reduction is often observed, which enables tumor cell immune escape, resulting in tumor progression and tolerance to immunotherapy (36).
Autophagy has a dual role in antigen presentation, the key process of anti-cancer immune responses. Activation of autophagy promotes antigen presentation to CD8+T lymphocytes mediated by DCs, which then stimulates cytotoxic responses (37,38). Besides, autophagy can also disrupt the process of antigen presentation. As an autophagy cargo receptor gene, NBR1 targets and degrades the MHC I, thus disrupting its antigen presentation ability to CD8+ T cells, which in turn can be reversed by autophagy inhibitor (39). Meanwhile, mucin domain protein-4 (TIM-4) binds to AMPK-a1 and activates the autophagy, and degrades the TAA, thus disrupting the antigen presentation and leading to the reduction of CD8+ T cells (40).
TME, which consists of the tumor and other cells (stromal cells, extracellular matrix (ECM), and immune cells, such as natural killer (NK) cells, tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), and T and B lymphocytes), is essential for the progression, maintenance, metastasis, and response to therapy of different types of cancers (41). Previous studies have shown that the immunosuppressive compartments of TME, including the autophagy activity of MDSCs, M2 macrophage polarization, and T cells regulatory (Tregs) recruitment, are associated with immune evasion (13,42,43). In our study, CD8 T cells and DCs had higher, while the M2 macrophage had lower infiltrations in the low-risk groups. Therefore, we reasonably speculated that IArlncRNAs are directly or indirectly involved in the immune regulation of TME, as well as autophagy.
This study has a few limitations. First, the data obtained from public databases were limited, and the clinical data were not comprehensive, which may lead to potential errors or deviations. Second, in this study, we did not verify the stability of IArlncRNAs prognostic characteristics in another independent data set and further experiments. Third, all the data used in this study were obtained from Western countries; therefore, careful consideration should be taken when applying our conclusions to patients from Asian countries.
Conclusions
In this study, we constructed a prognostic model with excellent predictive ability based on 4 IArlncRNAs in patients with bladder cancer. IArlncRNAs are critical factors, directly or indirectly involved in the immune regulation of TME and autophagy.
Acknowledgments
Funding: This study is supported by the High-level Hospital Construction Research Project of Maoming People’s Hospital and Guangdong Medical Science and Technology Research Fund (NO. 2017101819316701).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-560
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-560). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur Urol 2017;71:96-108. [Crossref] [PubMed]
- Zang Y, Li X, Cheng Y, et al. An overview of patients with urothelial bladder cancer over the past two decades: a Surveillance, Epidemiology, and End Results (SEER) study. Ann Transl Med 2020;8:1587. [Crossref] [PubMed]
- DeGeorge KC, Holt HR, Hodges SC. Bladder Cancer: Diagnosis and Treatment. Am Fam Physician 2017;96:507-14. [PubMed]
- Suh J, Yoo S. Role of immunotherapy in Bacillus Calmette-Guérin unresponsive: non-muscle invasive bladder cancer. Transl Cancer Res 2020;9:6537-45. [Crossref]
- Fonteyne V, Ost P, Bellmunt J, et al. Curative Treatment for Muscle Invasive Bladder Cancer in Elderly Patients: A Systematic Review. Eur Urol 2018;73:40-50. [Crossref] [PubMed]
- Kluth LA, Black PC, Bochner BH, et al. Prognostic and Prediction Tools in Bladder Cancer: A Comprehensive Review of the Literature. Eur Urol 2015;68:238-53. [Crossref] [PubMed]
- Sim KC, Sung DJ. Role of magnetic resonance imaging in tumor staging and follow-up for bladder cancer. Transl Androl Urol 2020;9:2890-907. [Crossref] [PubMed]
- Roupret M, Babjuk M, Comperat E, et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2017 Update. Eur Urol 2018;73:111-22. [Crossref] [PubMed]
- Zehnder P, Studer UE, Daneshmand S, et al. Outcomes of radical cystectomy with extended lymphadenectomy alone in patients with lymph node-positive bladder cancer who are unfit for or who decline adjuvant chemotherapy. BJU Int 2014;113:554-60. [Crossref] [PubMed]
- Jiang A, Liu N, Bai S, et al. The Construction and Analysis of Tumor-Infiltrating Immune Cells and ceRNA Networks in Bladder Cancer. Front Genet 2020;11:605767 [Crossref] [PubMed]
- Zheng Z, Mao S, Zhang W, et al. Dysregulation of the Immune Microenvironment Contributes to Malignant Progression and Has Prognostic Value in Bladder Cancer. Front Oncol 2020;10:542492 [Crossref] [PubMed]
- Qing L, Gu P, Liu M, et al. Extracellular Matrix-Related Six-lncRNA Signature as a Novel Prognostic Biomarker for Bladder Cancer. Onco Targets Ther 2020;13:12521-38. [Crossref] [PubMed]
- Cunha LD, Yang M, Carter R, et al. LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 2018;175:429-41 e16. [Crossref] [PubMed]
- He RQ, Zhou XG, Yi QY, et al. Prognostic Signature of Alternative Splicing Events in Bladder Urothelial Carcinoma Based on Spliceseq Data from 317 Cases. Cell Physiol Biochem 2018;48:1355-68. [Crossref] [PubMed]
- Wu X, Lv D, Cai C, et al. A TP53-Associated Immune Prognostic Signature for the Prediction of Overall Survival and Therapeutic Responses in Muscle-Invasive Bladder Cancer. Front Immunol 2020;11:590618 [Crossref] [PubMed]
- Li H, Lu H, Cui W, Huang Y, et al. A TP53-based immune prognostic model for muscle-invasive bladder cancer. Aging (Albany NY) 2020;13:1929-46. [Crossref] [PubMed]
- Mou Z, Yang C, Zhang Z, et al. Transcriptomic Analysis of Glycolysis-Related Genes Reveals an Independent Signature of Bladder Carcinoma. Front Genet 2020;11:566918 [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Zhang Q, Su M, Lu G, et al. The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol Cancer 2013;12:101. [Crossref] [PubMed]
- Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016;539:452-5. [Crossref] [PubMed]
- Guo H, Ahmed M, Zhang F, et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat Genet 2016;48:1142-50. [Crossref] [PubMed]
- Miyamoto DT, Mouw KW, Feng FY, et al. Molecular biomarkers in bladder preservation therapy for muscle-invasive bladder cancer. Lancet Oncol 2018;19:e683-e95. [Crossref] [PubMed]
- Quinn JJ, Ilik IA, Qu K, et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol 2014;32:933-40. [Crossref] [PubMed]
- Yan X, Hu Z, Feng Y, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015;28:529-40. [Crossref] [PubMed]
- Boon RA, Jae N, Holdt L, et al. Long Noncoding RNAs: From Clinical Genetics to Therapeutic Targets? J Am Coll Cardiol 2016;67:1214-26. [Crossref] [PubMed]
- Choudhari R, Sedano MJ, Harrison AL, et al. Long noncoding RNAs in cancer: From discovery to therapeutic targets. Adv Clin Chem 2020;95:105-47. [Crossref] [PubMed]
- Dyrskjot L, Reinert T, Algaba F, et al. Prognostic Impact of a 12-gene Progression Score in Non-muscle-invasive Bladder Cancer: A Prospective Multicentre Validation Study. Eur Urol 2017;72:461-9. [Crossref] [PubMed]
- Wang W, Zhao Z, Yang F, et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J Neurooncol 2018;136:263-71. [Crossref] [PubMed]
- Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol 2014;15:R47. [Crossref] [PubMed]
- Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9:e107468 [Crossref] [PubMed]
- Zhan Y, Chen Z, He S, et al. Long non-coding RNA SOX2OT promotes the stemness phenotype of bladder cancer cells by modulating SOX2. Mol Cancer 2020;19:25. [Crossref] [PubMed]
- Zheng R, Du M, Wang X, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer 2018;17:143. [Crossref] [PubMed]
- Zhan Y, Du L, Wang L, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer 2018;17:142. [Crossref] [PubMed]
- Munz C. Autophagy Beyond Intracellular MHC Class II Antigen Presentation. Trends Immunol 2016;37:755-63. [Crossref] [PubMed]
- Munz C. Enhancing immunity through autophagy. Annu Rev Immunol 2009;27:423-49. [Crossref] [PubMed]
- Reeves E, James E. Antigen processing and immune regulation in the response to tumours. Immunology 2017;150:16-24. [Crossref] [PubMed]
- Wang Y, Lin YX, Wang J, et al. In Situ Manipulation of Dendritic Cells by an Autophagy-Regulative Nanoactivator Enables Effective Cancer Immunotherapy. ACS Nano 2019;13:7568-77. [Crossref] [PubMed]
- Li Y, Hahn T, Garrison K, et al. The vitamin E analogue alpha-TEA stimulates tumor autophagy and enhances antigen cross-presentation. Cancer Res 2012;72:3535-45. [Crossref] [PubMed]
- Yamamoto K, Venida A, Yano J, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020;581:100-5. [Crossref] [PubMed]
- Baghdadi M, Yoneda A, Yamashina T, et al. TIM-4 glycoprotein-mediated degradation of dying tumor cells by autophagy leads to reduced antigen presentation and increased immune tolerance. Immunity 2013;39:1070-81. [Crossref] [PubMed]
- Roma-Rodrigues C, Mendes R, Baptista PV, et al. Targeting Tumor Microenvironment for Cancer Therapy. Int J Mol Sci 2019;20:840. [Crossref] [PubMed]
- Alissafi T, Hatzioannou A, Mintzas K, et al. Autophagy orchestrates the regulatory program of tumor-associated myeloid-derived suppressor cells. J Clin Invest 2018;128:3840-52. [Crossref] [PubMed]
- Rao S, Tortola L, Perlot T, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun 2014;5:3056. [Crossref] [PubMed]
(English Language Editors: J. Collie and J. Gray)


