
Trends in urine biomarker discovery for urothelial bladder cancer: DNA, RNA, or protein?
Introduction
Urothelial bladder cancer (UBC) typically presents as painless haematuria, demanding prompt investigation by cystoscopy and upper tract imaging to ascertain diagnosis (1). Non-muscle-invasive bladder cancer (NMIBC, stages: Tis/Ta/T1) comprises 75–80% of new UBC diagnoses, with muscle-invasive disease (MIBC, stage: T2+) comprising the remainder (2). Following initial transurethral resection of tumour (TURBT) and adjuvant intravesical therapy where indicated, long-term surveillance via serial cystoscopy remains an essential mainstay of NMIBC management due to the high incidence of recurrence and a risk of progression to MIBC (1,3,4). Despite cystoscopy being invasive, uncomfortable, expensive and incurring morbidity, no alternative is recommended in current clinical guidelines (1,5).
Following TURBT, tumour grade, TNM stage and other clinico-pathological parameters are used to classify disease according to prognosis and to tailor treatment (1),but there exists significant heterogeneity within these ‘disease states’ limiting the applicability of existing classifiers. Given the complex nature of UBC, this is not unexpected; hence, understanding the underlying biology of urothelial carcinogenesis could substantially transform clinical efficiency and patient management. The molecular pathways in bladder cancer, highlighting key pathways of hallmark features leading to urothelial transformation were described in 2005 in the ‘pre-genomics’ era (6,7), and the more aggressive nature of tumours derived from the basal layer of the urothelium was first considered as early as 2008 (8,9). More recently, DNA and RNA sequencing-based analyses have significantly expanded and detailed these concepts (10-14), with Kamoun et al. outlining a consensus molecular classification of MIBC based upon 6 molecular subtypes and associated clinical outcomes (15). Hence, ongoing technological developments in “omics” with concomitant verification via bioinformatics have opened a plethora of possibilities for potential biomarkers. A key output of such research would be the identification of diagnostic molecular markers measurable in urine with high specificity and sensitivity in order to streamline UBC diagnosis and NMIBC surveillance, aid clinical decision-making, and improve cost-effectiveness.
Cystoscopy has been considered as the ‘gold standard’ for the diagnosis of bladder cancer since its application within an outpatient setting in London in 1984 (16). Subsequent improvements have been made to the method to assist in diagnosis and surveillance viz. blue-light cystoscopy (photodynamic diagnosis) and narrow-band imaging (17-20). Although highly operator-dependent, the overall sensitivity surpasses any other current form of diagnostic modality for the detection of UBC with sensitivity estimated at 85%, with 87% specificity (21).
Novel biomarker discovery has been fuelled by exhaustive research efforts in the last few decades to determine an optimal molecular substrate. A number of urine-based tests have been approved by the US FDA (e.g., NMP22, NMP22 BladderChek Test, BTA TRAK, BTA Stat, UroVysion® and ImmunoCyt), but none are robust enough to facilitate early detection and risk stratification of UBC for routine clinical implementation (22). Instead, urinary cytology remains the most commonly used adjunct to cystoscopy, with high sensitivity to diagnose high-grade disease (sensitivity of 84%), despite its limitations in identifying low-risk cancer (sensitivity 16–48%) (23).
Conceptually, a biomarker would identify targets in the DNA, RNA or protein of tumour cells and be able to distinguish them from normal cells. As urine is in direct contact with the tumour tissue itself, appreciable amounts of protein and genomic material can be found within urine, either in exfoliated UBC cells or as cell-free constituents. Sample collection is inexpensive, painless, and repeatable, and so urine is a logical source for biomarker analysis—a ‘liquid biopsy’, a simple example of which includes voided urinary cytopathology (VUC) where cells are directly observed microscopically.
In this current review, we have outlined the changing trends over the last five decades in the pursuit of an optimal diagnostic urinary biomarker, focusing on the different classes of biomolecule that can act as a biomarker substrate for UBC- DNA, RNA, protein.
Urine biomarker trends
Searches were performed on Medline and PubMed platforms for available literature using the following keywords: (“urinary bladder neoplasms”[MeSH Terms] OR (“urinary”[All Fields] AND “bladder”[All Fields] AND “neoplasms”[All Fields]) OR “urinary bladder neoplasms”[All Fields] OR (“bladder”[All Fields] AND “cancer”[All Fields]) OR “bladder cancer”[All Fields]) OR “urine”[MeSH Subheading] OR “urine”[All Fields] OR “urine”[MeSH Terms] OR “urines”[All Fields]) AND (“biomarker s”[All Fields] OR “biomarkers”[MeSH Terms] OR “biomarkers”[All Fields] OR “biomarker”[All Fields]). Articles from January 1970 to January 2020 were included with the prerequisites of being in the English language, with available abstracts. Review articles, letters, editorials, or comments were excluded. Journal articles were included if the reported studies were performed on urine. Biomarkers were categorized as DNA/genomic, RNA/transcriptomic or protein.
Data were collated from articles regarding: the biomarker assay type, the technique utilized for analysis, assessment utility, and where available—associated sensitivity, specificity, positive predictive value and negative predictive value.
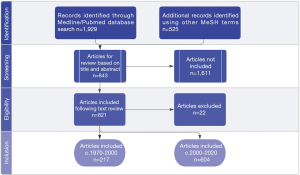
The database search identified 1929 articles. Two additional searches were created using the following: “urine biomarker DNA” and “urine biomarker RNA”, resulting in 301 and 224 results, respectively. 843 articles were reviewed (Figure 1) that reported either DNA, RNA or protein biomarkers (Figure 2A), with a breakdown of classes of RNA or DNA marker and their discovery observed over 5 decades (Figure 2B).
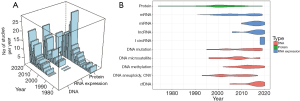
Proteins
Early protein biomarker research studies were limited to antibody-based measurements of one, or a small number of, biomarker candidates e.g., Carcinoembryonic antigen (CEA) (24), tissue plasminogen activator (TPA) (25), fibronectin (26). The advent of “proteomics” (2D-electrophoresis in the 1970s and protein identification by mass spectrometry in the 1980s) enabled biomarker discovery via comparison of the protein composition of cell lines and tumour and non-tumour tissues and urine e.g., (27,28). Since 2000 there have been enormous improvements in the speed, accuracy and sensitivity of mass spectrometers and the combination of two-dimensional fractionation of tryptic peptides with mass spectrometry (or “shotgun proteomics”) has enabled more in-depth proteomic analysis allowing, for example, identification and quantitation of >1,500 proteins in human urine (29,30). In the last decade targeted peptide mass spectrometry (“MRM”) and multi-plex immunoassay platforms have enabled the measurement of panels of protein biomarker candidates derived from transcriptomic and proteomic analyses (31).
From the 1970s to 2000s, 208 studies evaluated the utility of urinary proteins as biomarkers. We found 25 studies investigating the potential use of urinary CEA as an adjunct to cystoscopy or as a stand-alone diagnostic biomarker (24,32-55). Twelve studies investigated survivin, a protein which inhibits apoptosis allowing tumour cell survival (56-68). Fragments of cellular cytoskeletal proteins cytokeratins 8, 18, 19 and 20 (69) have also been found in urine of UBC patients, leading to the development of Urothelial Bladder Carcinoma- antigen assay (UBC) (66,70-79) and Cyfra21.1 (80-91). Vascular endothelial growth factor (VEGF) has been investigated in 10 studies (59,64,86,92-98). Several proteins have been commercialised as bladder cancer detection tests (Figure 3) although only NMP22 and BTA measured as soluble proteins in urine, and immunocyte which measures mucins and a glycoform of CEA on exfoliated cells have gained FDA approval. NMP22 and BTA can be measured quantitatively using Enzyme linked immunosorbent assay (ELISAs) or qualitative point-of-care tests, validated in 117, 97 and 43 studies, respectively.
Combining protein biomarkers into multi-analyte tests has largely failed to increase sensitivity and specificity although it remains possible that tumour specific variants of proteins (e.g., novel splice variants or glycoforms) could offer improved performance. Though a large proportion of bladder cancer biomarker discovery research has focused on proteins, developments in genomics, epigenetics and transcriptomics have shifted the research towards nucleic acid biomarkers (discussed below). This is evident in the continued decline in the protein biomarker studies reported in the last decade.
DNA and genomics
Game changing technological developments such as polymerase chain reaction (PCR) in the 1970s, microarrays in the 1980s and massively parallel sequencing (next generation sequencing or NGS) in the 2000s have facilitated biomarker research, characterization of cellular and molecular pathways, and provided targets for novel therapeutics and drug discovery (10,11). The concept of “liquid biopsy” fostered the analysis of DNA in plasma and urine, in studies involving methylation, mutation, and copy number variations (CNV) (111). Due to the molecular heterogeneity of UBC, multiplexed genomic profiling of DNA is likely to prove more useful than individual markers.
DNA: copy number changes and microsatellite instability
Loss of chromosome 9p, 9q and 17 (112,113) are common early events in bladder cancer and a large fraction of the entire genome undergoes copy number changes in high-grade disease (114-116). These phenomena are utilised by several commercially available bladder cancer tests; UroVysion uses fluorescence in situ hybridization (FISH) to detect aneuploidy for chromosomes 3, 7, 17, and loss of 9p21 in exfoliated cells (117-122) and BCA-1 uses array comparative genomic hybridisation to analyse genome wide copy number changes (109,123,124) (Figure 3). More recently, microarrays and next generation sequencing have been used to study copy number changes in urine cell-free and pellet DNA (116,125-127). In the 1990s microsatellite instability markers were widely researched for diagnosis but this has now ceased (128,129).
DNA: mutations
Tumour DNA from urine can be extracted following centrifugation (cell pellet DNA, cpDNA) or as fragmented cell-free DNA (cfDNA) in the supernatant (130,131); both forms are admixed with a large background of normal DNA. However, using highly sensitive analytical approaches such as digital droplet PCR (ddPCR) and next-generation sequencing (NGS), it is possible to detect cancer-associated mutations at very low levels in urine.
Two distinct pathways have been recognized that encourage the development of NMIBC or MIBC, by activation of MAPK-PIK3 pathway or disruption of cell cycle regulators (132,133). 11 studies have found mutations in FGFR3, RAS, PIK3CA to translate phenotypically to NMIBC (99,134-144), whereas more aggressive MIBC states are generally characterized by mutations in TP53, CDKN2A, MLL and ERBB2 (n=6 studies) (12,145-150). 11 studies were found on TERT promoter mutation; which has been observed in the early stages of tumour development occurring in 74% of incident and recurrent cases, and is also independently related to poor survival outcomes reflecting their potential usage as prognostic markers (151-155). The analyses of multiple DNA mutations (19 studies) have provided better overall sensitivity and specificity for the detection of NMIBC by use of next generation sequencing (99,115,135,141-145,149,156-159).
DNA: methylation
DNA methylation status can be readily assessed in bisulphite converted cpDNA and cfDNA through quantitative methylation specific PCR (QMSP-PCR), pyrosequencing, NGS and/or ddPCR (Table 1). We found 54 studies that investigated methylation markers in urine for UBC detection. An assay based on 15 methylation markers has been commercialised (Epicheck) and another using a combination of 3 DNA methylation (OTX1, ONECUT2, TWIST1) and 3 mutation markers (HRAS, FGFR3, TERT)- Assure MDx functional biomarker (Figure 3).
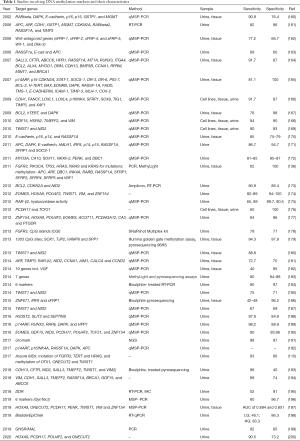
Full table
RNA and transcriptomics
Expression profiling with microarrays studies and more recently RNA sequencing has identified numerous overexpressed transcripts in bladder cancer. Most of this work focused on protein coding messenger (m)RNAs although additional classes of RNA [micro(mi)RNA, long non-coding (lnc)RNA and circular (circ)RNA] have come to the fore recently (described below). We found 61 urine mRNA biomarker studies, mostly using RT-qPCR to measure specific gene products. The commercially available Cx bladder test analyses a panel of 5 genes that are overexpressed in bladder cancer by RT-qPCR.
miRNA
Micro RNAs are short non-coding RNAs that impose post-translational effects by binding onto the 3’ untranslated region (UTR) of their target mRNA, to regulate gene expression. miRNA-altered gene expression has been found to initiate and promote tumour progression, making them potential diagnostic and prognostic indicators. MiRNAs are stable and detectable in body fluids, including urine (201,202). With the aid of computational biological methods, identification and stratification of miRNA transcripts can be scrutinized for expression levels. Subsequent analysis can be validated using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), ddPCR, microarray, pyrosequencing or NGS. Since their discovery (203), multiple studies have been carried out to research this entity in urine (Table 2).
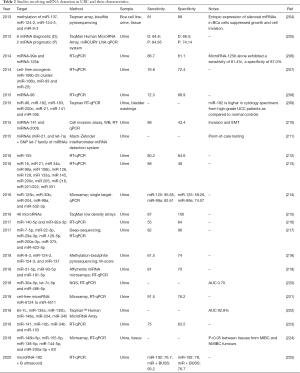
Full table
lncRNA
LncRNAs are long RNA molecules that do not encode proteins. They may serve as regulators of gene expression and recent high throughput profiling studies have shown dysregulation of lncRNAs to promote oncogenesis. lncRNAs found in urine have been associated with UBC (Table 3) which merits further exploration to uncover their functional roles in tumorigenesis. Urothelial-carcinoma-associated 1 (UCA-1) lncRNA is a potential biomarker that is upregulated in the presence of UBC in proportion to the grade of disease (226,233).
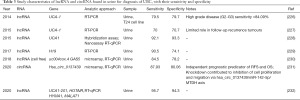
Full table
circRNA
An emerging novel class of RNAs are circular RNAs (circRNA). These are single-stranded RNAs that are covalently linked through backsplicing events to form a closed continuous loop. They are stable, immune to degradation by RNAases, and can be found in urine (Table 3) (234). Emerging evidence indicates their role in regulating gene expression by “sponging” miRNAs (235). Recent studies profiling circRNA in tumour tissue (236), demonstrate that their relative expression levels are indicative of tumour grade and stage (231,237), and involvement in urothelial carcinogenesis (238), providing insights into a fascinating new genre of biomarkers.
Conclusion and future prospects
Over the course of 5 decades, technological advances have transformed biomarker discovery from immunoassays for single protein biomarkers through to whole genome sequencing and accessing new classes of modifications and biomolecules. Challenges remain to detect even the slightest amount of tumour DNA from large amount of background normal DNA. Whereas the prospects of available urinary biomarkers are generally promising, an element of bias in regard to sample selection still exists i.e., performance evaluation of the test relies on the prevalence of underlying disease in specific cohorts. There may also be resistance from medical practitioners to change practice until biomarker tests match or exceed the performance of cystoscopy (even though cystoscopy itself is imperfect).
Additionally, bladder cancer research itself faces significant bias in terms of funding opportunities. It has been reported that in 2017/18, £4.5 million were spent on bladder cancer by Cancer Research UK, compared to £22.1 million for prostate cancer and £7.3 million for renal cancer (239). Many biomarkers exist with the potential to transform bladder cancer detection and even provide personalized medicine, but due to poor research funding and lack of level I evidence, uptake into clinical practice is hesitant.
Large-scale multicentre prospective clinical trials are required to validate the performance of promising biomarkers that have demonstrated potential clinical utility: sensitivities and specificities to match or exceed cystoscopy, the capability to be multiplexed with high throughput, and accompanied by limited impact on resources (22). Given the funding challenges across all of bladder cancer research, such studies have not yet been undertaken. Hence, the current generation of highly promising biomarkers remain ‘on the bench’, both physically and metaphorically. Notwithstanding, a role of cystoscopy will be ever present for the diagnosis of other conditions of the lower urinary tract.
As we enter the 2020s, further diagnostic biomarker classes are emerging. For example, the profiling of the urinary bladder microbiome to investigate causal relationships with UBC. In other cancer settings, the microbiome possesses mutagenic properties that can initiate tumorigenesis by way of chronic insult, such as in the case of Helicobacter pylori infection and gastric cancer (240).Recent studies have reported an abundance of Acinetobacter spp. from patients with HR-NMIBC (241-243). The preliminary studies suggest that there is potential in investigating the urinary microbiome for early screening of UBC, but much remains to be discovered.
Likewise, extracellular vesicles (EV)—small spherical membrane vesicles secreted by all cells, cargo nucleic acids or proteins, and are the responsible machinery for intercellular communication (244). They are especially important in cell-cell signalling, extracellular-matrix (ECM) remodelling, and distant spread/ metastasis when secreted by cancer cells as exosomes (245). Exosomes act as vehicles for the transport of mRNA, miRNA, lncRNA, tumour DNA and protein to build and nurture tumour micro-environment (TME) for development and progression of cancer. EVs have been isolated from urine supernatant through ultracentrifugation and characterized by MS (246), nanoparticle tracking (247), immunoaffinity-capture, microfluidic-filtration (248). Whichever method is utilized, the challenge is to obtain sufficient quantity of undiluted EV for analysis, hence there is no consensus on one set technique. EVs have gained attention in the second-half (2015–2020) of the last decade and following a standardized technique for analysis, they could be revolutionary for the forthcoming era.
And so, having demonstrated the evolution of research into the biomolecule substrates for urinary diagnostic biomarkers from the 1970s to date, from proteins to DNA/RNA, the field continues to expand. Notwithstanding, clinically relevant and acceptable sensitivities and specificities at affordable costs from the current generation of diagnostic urinary biomarkers are very much within reach.
Acknowledgments
Richard T. Bryan has contributed to advisory boards for Olympus Medical Systems with regards to narrow band imaging cystoscopy.
Funding: Nada Humayun-Zakaria’s research is funded by QEHB Charities.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Mieke Van Hemelrijck and Netty Kinsella) for the series “Expectant Management in Genitourinary Malignancies (Prostate, Bladder, Kidney)” published in Translational Andrology and Urology. The article has undergone external peer review.
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1327). The series “Expectant Management in Genitourinary Malignancies (Prostate, Bladder, Kidney)” was commissioned by the editorial office without any funding or sponsorship. Dr. RTB reports other from Janssen EMEA, grants from UroGen Pharma, grants from QED Therapeutics, outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Babjuk M, Burger M, Compérat EM, et al. European Association of Urology Guidelines on Non-muscle-invasive Bladder Cancer (TaT1 and Carcinoma In Situ) - 2019 Update. Eur Urol 2019;76:639-57. [Crossref] [PubMed]
- Bryan RT, Zeegers MP, van Roekel EH, et al. A comparison of patient and tumour characteristics in two UK bladder cancer cohorts separated by 20 years. BJU Int 2013;112:169-75. [Crossref] [PubMed]
- Cambier S, Sylvester RJ, Collette L, et al. EORTC Nomograms and Risk Groups for Predicting Re-currence, Progression, and Disease-specific and Overall Survival in Non–Muscle-invasive Stage Ta–T1 Urothelial Bladder Cancer Patients Treated with 1–3 Years of Maintenance Bacillus Calmette-Guérin. Eur Urol 2016;69:60-9. [Crossref] [PubMed]
- Jobczyk M, Stawiski K, Fendler W, et al. Validation of EORTC, CUETO, and EAU risk stratification in prediction of recurrence, progression, and death of patients with initially non-muscle-invasive bladder cancer (NMIBC): A cohort analysis. Cancer Med 2020;9:4014-25. [Crossref] [PubMed]
- Svatek RS, Hollenbeck BK, Holmäng S, et al. The economics of bladder cancer: costs and considerations of caring for this disease. Eur Urol 2014;66:253-62. [Crossref] [PubMed]
- Bryan RT, Hussain SA, James ND, et al. Molecular pathways in bladder cancer: part 1. BJU Int 2005;95:485-90. [Crossref] [PubMed]
- Bryan RT, Hussain SA, James ND, et al. Molecular pathways in bladder cancer: Part 2. BJU Int 2005;95:491-6. [Crossref] [PubMed]
- Bryan RT, Atherfold PA, Yeo Y, et al. Cadherin switching dictates the biology of transitional cell car-cinoma of the bladder: ex vivo and in vitro studies. J Pathol 2008;215:184-94. [Crossref] [PubMed]
- Bryan RT, Tselepis C. Cadherin switching and bladder cancer. J Urol 2010;184:423-31. [Crossref] [PubMed]
- Hurst CD, Platt FM, Taylor CF, et al. Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis. Clin Cancer Res 2012;18:5865-77. [Crossref] [PubMed]
- Sjödahl G, Lauss M, Lövgren K, et al. A Molecular Taxonomy for Urothelial Carcinoma. Clin Cancer Res 2012;18:3377-86. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016;30:27-42. [Crossref] [PubMed]
- Czerniak B, Dinney C, McConkey D. Origins of Bladder Cancer. Annu Rev Pathol 2016;11:149-74. [Crossref] [PubMed]
- Kamoun A, de Reyniès A, Allory Y, et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur Urol 2020;77:420-33. [Crossref] [PubMed]
- Fowler CG. Fibrescope urethrocystoscopy. Br J Urol 1984;56:304-7. [Crossref] [PubMed]
- Tschirdewahn S, Harke NN, Hirner L, et al. Narrow-band imaging assisted cystoscopy in the follow-up of patients with transitional cell carcinoma of the bladder: a randomized study in comparison with white light cystoscopy. World J Urol 2020;38:1509-15. [Crossref] [PubMed]
- Drejer D, Moltke AL, Nielsen AM, et al. DaBlaCa-11: Photodynamic Diagnosis in Flexible Cystos-copy-A Randomized Study With Focus on Recurrence. Urology 2020;137:91-6. [Crossref] [PubMed]
- Lotan Y, Chaplin I, Ahmadi H, et al. Prospective evaluation of blue-light flexible cystoscopy with hex-aminolevulinate in non-muscle-invasive bladder cancer. BJU Int 2021;127:108-13. [Crossref] [PubMed]
- Bryan RT, Billingham LJ, Wallace DM. Narrow-band imaging flexible cystoscopy in the detection of recurrent urothelial cancer of the bladder. BJU Int 2008;101:702-5; discussion 705-6. [Crossref] [PubMed]
- Zheng C, Lv Y, Zhong Q, et al. Narrow band imaging diagnosis of bladder cancer: systematic review and meta-analysis. BJU Int 2012;110:E680-7. [Crossref] [PubMed]
- Schmitz-Dräger BJ, Droller M, Lokeshwar VB, et al. Molecular markers for bladder cancer screening, early diagnosis, and surveillance: the WHO/ICUD consensus. Urol Int 2015;94:1-24. [Crossref] [PubMed]
- Yafi FA, Brimo F, Steinberg J, et al. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol Oncol 2015;33:66.e25-31. [Crossref] [PubMed]
- Nery R, Barsoum AL, Bullman H, et al. Carcinoembryonic antigen-like substances of human urothelial carcinomas. Isolation of components from pathological urine and comparison with colorectal carcinoma antigens. Biochem J 1974;139:431-40. [Crossref] [PubMed]
- Carbin BE, Collins VP, Ekman P. Tissue polypeptide antigen (TPA), some cytokeratins and epithelial membrane antigen (EMA) in normal, inflamed and malignant urothelium. Urol Res 1987;15:191-4. [Crossref] [PubMed]
- Malmström PU, Larsson A, Johansson S. Urinary fibronectin in diagnosis and follow-up of patients with urinary bladder cancer. Br J Urol 1993;72:307-10. [Crossref] [PubMed]
- Yang G, Xu Z, Lu W, et al. Quantitative Analysis of Differential Proteome Expression in Bladder Cancer vs. Normal Bladder Cells Using SILAC Method. PLoS One 2015;10:e0134727 [Crossref] [PubMed]
- Shimwell NJ, Bryan RT, Wei W, et al. Combined proteome and transcriptome analyses for the discovery of urinary biomarkers for urothelial carcinoma. Br J Cancer 2013;108:1854-61. [Crossref] [PubMed]
- Di Meo A, Batruch I, Yousef AG, et al. An integrated proteomic and peptidomic assessment of the normal human urinome. Clin Chem Lab Med 2017;55:237-47. [Crossref] [PubMed]
- Zhang Q, Huang S, Luo H, et al. Eight-plex iTRAQ labeling and quantitative proteomic analysis for human bladder cancer. Am J Cancer Res 2017;7:935-45. [PubMed]
- Duriez E, Masselon CD, Mesmin C, et al. Large-Scale SRM Screen of Urothelial Bladder Cancer Candidate Biomarkers in Urine. J Proteome Res 2017;16:1617-31. [Crossref] [PubMed]
- Nery R, James R, Barsoum AL, et al. Isolation and partial characterization of macromolecular urinary aggregates containing carcinoembryonic antigen-like activity. Br J Cancer 1974;29:413-24. [Crossref] [PubMed]
- Guinan P, Dubin A, Bush I, et al. The CEA test in urologic cancer: an evaluation and a review. Oncology 1975;32:158-68. [Crossref] [PubMed]
- Coombers GB, Hall RR, Laurence JR, et al. Urinary carcinoembryonic antigen (CEA)-like molecules and urothelial malignancy: a clinical appraisal. Br J Cancer 1975;31:135-42. [Crossref] [PubMed]
- Monaco AP, Gozzo JJ, Schlesinger RM, et al. Immunological detection of human bladder carcinoma. Ann Surg 1975;182:325-33. [Crossref] [PubMed]
- Wahren B, Edsmyr F, Zimmerman R. Measurement of urinary CEA-like substance. An aid in man-agement of patients with bladder carcinoma. Cancer 1975;36:1490-5. [Crossref] [PubMed]
- Korsten CB, Persijn JP, Renaud J, et al. Carcino-embryonic antigen activity in urine of patients with bladder carcinoma. Clinical evaluation of carcino-embryonic antigen. II. J Clin Chem Clin Biochem 1976;14:389-93. [PubMed]
- Persijn JP, Korsten CB, Batterman JJ, et al. Clinical significance of urinary carcino-embryonic antigen estimations during the follow-up of patients with bladder carcinoma or previous bladder carcinoma. Clinical evaluation of carcino-embryonic antigen, III. J Clin Chem Clin Biochem 1976;14:395-9. [Crossref] [PubMed]
- Wahren B, Edsmyr F. Carcinoembryonic antigen in serum, urine and cells of patients with bladder carcinoma. Urol Res 1978;6:221-4. [Crossref] [PubMed]
- Guinan PD, McKiel C, Sundar B, et al. The carcinoembryonic antigen test in urologic cancer. Natl Cancer Inst Monogr 1978;32:225-9. [PubMed]
- Colleen S, Ek A, Gullberg B, et al. Carcinoembryonic antigen in urine in patients with urothelial carci-noma. An expression for the extent of inflammatory reaction of the urinary tract. Scand J Urol Nephrol 1979;13:149-53. [Crossref] [PubMed]
- Glashan RW, Higgins E, Neville AM. The clinical value of plasma and urinary carcinoembryonic antigen (CEA) assays in patients with haematuria and urothelial carcinoma. Eur Urol 1980;6:344-6. [Crossref] [PubMed]
- James K, Alroy J, Miller AW, et al. Effects of therapy on carcinoembryonic antigen activity in the urines of patients with cancer of the bladder. Am J Clin Pathol 1980;73:250-3. [Crossref] [PubMed]
- Zimmerman R, Wahren B, Edsmyr F. Assessment of serial CEA determinations in urine of patients with bladder carcinoma. Cancer 1980;46:1802-9. [Crossref] [PubMed]
- Shevchuk MM, Fenoglio CM, Richart RM. Carcinoembryonic antigen localization in benign and ma-lignant transitional epithelium. Cancer 1981;47:899-905. [Crossref] [PubMed]
- Wahren B, Nilsson B, Zimmerman R. Urinary CEA for prediction of survival time and recurrence in bladder cancer. Cancer 1982;50:139-45. [Crossref] [PubMed]
- Huland H, Otto U, Droese M. The value of urinary cytology, serum and urinary carcinoembryonic antigen, rheumatoid factors, and urinary immunoglobulin concentration as tumor markers or prognostic factors in predicting progression of superficial bladder cancer. Eur Urol 1983;9:346-9. [Crossref] [PubMed]
- Jakse G, Rauschmeier H, Rosmanith P, et al. Determination of carcinoembryonic antigen in tissue, serum and urine in patients with transitional cell carcinoma of the urinary bladder. Urol Int 1983;38:121-5. [Crossref] [PubMed]
- Tailly G, Cornelissen M, Vereecken RL, et al. Urinary carcinoembryogenic antigen (CEA) in the di-agnosis and follow-up of bladder carcinoma. Br J Urol 1983;55:501-7. [Crossref] [PubMed]
- Holmberg V, Nilsson B. Short-term prognosis of bladder cancer. Evaluation with urinary CEA. Anticancer Res 1984;4:121-2. [PubMed]
- Hetherington JW, Ewing R, Cooper EH. Evaluation of a double-monoclonal radioimmunoassay for the measurement of carcinoembryonic antigen in the urine of patients with bladder cancer. Eur Urol 1986;12:270-3. [Crossref] [PubMed]
- Tizzani A, Cassetta G, Cicigoi A, et al. Tumor markers (CEA, TPA and CA 19-9) in urine of bladder cancer patients. Int J Biol Markers 1987;2:121-4. [Crossref] [PubMed]
- Halim AB, el-Ahmady O, Hamza S, et al. Simultaneous determination of urinary CEA, ferritin and TPA in Egyptian bladder cancer patients. Int J Biol Markers 1992;7:234-9. [Crossref] [PubMed]
- Casetta G, Piana P, Cavallini A, et al. Urinary levels of tumour associated antigens (CA 19-9, TPA and CEA) in patients with neoplastic and non-neoplastic urothelial abnormalities. Br J Urol 1993;72:60-4. [Crossref] [PubMed]
- Fraser RA, Ravry MJ, Segura JW, et al. Clinical evaluation of urinary and serum carcinoembryonic antigen in bladder cancer. J Urol 1975;114:226-9. [Crossref] [PubMed]
- Smith SD, Wheeler MA, Plescia J, et al. Urine detection of survivin and diagnosis of bladder cancer. JAMA 2001;285:324-8. [Crossref] [PubMed]
- Ohsawa I, Nishimura T, Kondo Y, et al. Detection of urine survivin in 40 patients with bladder cancer. J Nippon Med Sch 2004;71:379-83. [Crossref] [PubMed]
- Davies B, Chen JJ, McMurry T, et al. Efficacy of BTA stat, cytology, and survivin in bladder cancer surveillance over 5 years in patients with spinal cord injury. Urology 2005;66:908-11. [Crossref] [PubMed]
- Sun Y, He DL, Ma Q, et al. Comparison of seven screening methods in the diagnosis of bladder cancer. Chin Med J (Engl) 2006;119:1763-71. [Crossref] [PubMed]
- Kenney DM, Geschwindt RD, Kary MR, et al. Detection of newly diagnosed bladder cancer, bladder cancer recurrence and bladder cancer in patients with hematuria using quantitative rt-PCR of urinary survivin. Tumour Biol 2007;28:57-62. [Crossref] [PubMed]
- Pu XY, Wang ZP, Chen YR, et al. The value of combined use of survivin, cytokeratin 20 and mucin 7 mRNA for bladder cancer detection in voided urine. J Cancer Res Clin Oncol 2008;134:659-65. [Crossref] [PubMed]
- Horstmann M, Bontrup H, Hennenlotter J, et al. Clinical experience with survivin as a biomarker for urothelial bladder cancer. World J Urol 2010;28:399-404. [Crossref] [PubMed]
- Eissa S, Shabayek MI, Ismail MF, et al. Diagnostic evaluation of apoptosis inhibitory gene and tissue inhibitor matrix metalloproteinase-2 in patients with bladder cancer. IUBMB Life 2010;62:394-9. [Crossref] [PubMed]
- Gogalic S, Sauer U, Doppler S, et al. Bladder cancer biomarker array to detect aberrant levels of proteins in urine. Analyst 2015;140:724-35. [Crossref] [PubMed]
- Chatziharalambous D, Lygirou V, Latosinska A, et al. Analytical Performance of ELISA Assays in Urine: One More Bottleneck towards Biomarker Validation and Clinical Implementation. PLoS One 2016;11:e0149471 [Crossref] [PubMed]
- Gleichenhagen J, Arndt C, Casjens S, et al. Evaluation of a New Survivin ELISA and UBC(®) Rapid for the Detection of Bladder Cancer in Urine. Int J Mol Sci 2018;19:226. [Crossref] [PubMed]
- Liang Z, Xin R, Yu Y, et al. Diagnostic value of urinary survivin as a biomarker for bladder cancer: a systematic review and meta-analysis of published studies. World J Urol 2018;36:1373-81. [Crossref] [PubMed]
- Yang Y, Xu J, Zhang Q. Detection of urinary survivin using a magnetic particles-based chemilumines-cence immunoassay for the preliminary diagnosis of bladder cancer and renal cell carcinoma combined with LAPTM4B. Oncol Lett 2018;15:7923-33. [PubMed]
- Morsi MI, Youssef AI, Hassouna ME, et al. Telomerase activity, cytokeratin 20 and cytokeratin 19 in urine cells of bladder cancer patients. J Egypt Natl Canc Inst 2006;18:82-92. [PubMed]
- Sánchez-Carbayo M, Herrero E, Megías J, et al. Initial evaluation of the new urinary bladder cancer rapid test in the detection of transitional cell carcinoma of the bladder. Urology 1999;54:656-61. [Crossref] [PubMed]
- Mian C, Lodde M, Haitel A, et al. Comparison of the monoclonal UBC-ELISA test and the NMP22 ELISA test for the detection of urothelial cell carcinoma of the bladder. Urology 2000;55:223-6. [Crossref] [PubMed]
- Heicappell R, Schostak M, Müller M, et al. Evaluation of urinary bladder cancer antigen as a marker for diagnosis of transitional cell carcinoma of the urinary bladder. Scand J Clin Lab Invest 2000;60:275-82. [Crossref] [PubMed]
- Mungan NA, Vriesema JL, Thomas CM, et al. Urinary bladder cancer test: a new urinary tumor marker in the follow-up of superficial bladder cancer. Urology 2000;56:787-92. [Crossref] [PubMed]
- Eissa S, Swellam M, Sadek M, et al. Comparative evaluation of the nuclear matrix protein, fibronectin, urinary bladder cancer antigen and voided urine cytology in the detection of bladder tumors. J Urol 2002;168:465-9. [Crossref] [PubMed]
- Hakenberg OW, Fuessel S, Richter K, et al. Qualitative and quantitative assessment of urinary cy-tokeratin 8 and 18 fragments compared with voided urine cytology in diagnosis of bladder carcinoma. Urology 2004;64:1121-6. [Crossref] [PubMed]
- Ecke TH, Arndt C, Stephan C, et al. Preliminary Results of a Multicentre Study of the UBC Rapid Test for Detection of Urinary Bladder Cancer. Anticancer Res 2015;35:2651-5. [PubMed]
- Ecke TH, Weiß S, Stephan C, et al. UBC(®) Rapid Test for detection of carcinoma in situ for bladder cancer. Tumour Biol 2017;39:1010428317701624 [Crossref] [PubMed]
- Pichler R, Tulchiner G, Fritz J, et al. Urinary UBC Rapid and NMP22 Test for Bladder Cancer Sur-veillance in Comparison to Urinary Cytology: Results from a Prospective Single-Center Study. Int J Med Sci 2017;14:811-9. [Crossref] [PubMed]
- Ecke TH, Weiß S, Stephan C, et al. UBC(®) Rapid Test-A Urinary Point-of-Care (POC) Assay for Diagnosis of Bladder Cancer with a focus on Non-Muscle Invasive High-Grade Tumors: Results of a Multicenter-Study. Int J Mol Sci 2018;19:3841. [Crossref] [PubMed]
- Dittadi R, Barioli P, Gion M, et al. Standardization of assay for cytokeratin-related tumor marker CYFRA21.1 in urine samples. Clin Chem 1996;42:1634-8. [Crossref] [PubMed]
- Senga Y, Kimura G, Hattori T, et al. Clinical evaluation of soluble cytokeratin 19 fragments (CYFRA 21-1) in serum and urine of patients with bladder cancer. Urology 1996;48:703-10. [Crossref] [PubMed]
- Sánchez-Carbayo M, Herrero E, Megías J, et al. Comparative sensitivity of urinary CYFRA 21-1, urinary bladder cancer antigen, tissue polypeptide antigen, tissue polypeptide antigen and NMP22 to detect bladder cancer. J Urol 1999;162:1951-6. [Crossref] [PubMed]
- Pariente JL, Bordenave L, Jacob F, et al. Analytical and prospective evaluation of urinary cytokeratin 19 fragment in bladder cancer. J Urol 2000;163:1116-9. [Crossref] [PubMed]
- Sánchez-Carbayo M, Urrutia M, Silva JM, et al. Comparative predictive values of urinary cytology, urinary bladder cancer antigen, CYFRA 21-1 and NMP22 for evaluating symptomatic patients at risk for bladder cancer. J Urol 2001;165:1462-7. [Crossref] [PubMed]
- Nisman B, Barak V, Shapiro A, et al. Evaluation of urine CYFRA 21-1 for the detection of primary and recurrent bladder carcinoma. Cancer 2002;94:2914-22. [Crossref] [PubMed]
- Bian W, Xu Z. Combined assay of CYFRA21-1, telomerase and vascular endothelial growth factor in the detection of bladder transitional cell carcinoma. Int J Urol 2007;14:108-11. [Crossref] [PubMed]
- Gkialas I, Papadopoulos G, Iordanidou L, et al. Evaluation of urine tumor-associated trypsin inhibitor, CYFRA 21-1, and urinary bladder cancer antigen for detection of high-grade bladder carcinoma. Urology 2008;72:1159-63. [Crossref] [PubMed]
- Huang YL, Chen J, Yan W, et al. Diagnostic accuracy of cytokeratin-19 fragment (CYFRA 21-1) for bladder cancer: a systematic review and meta-analysis. Tumour Biol 2015;36:3137-45. [Crossref] [PubMed]
- Kuang LI, Song WJ, Qing HM, et al. CYFRA21-1 levels could be a biomarker for bladder cancer: a meta-analysis. Genet Mol Res 2015;14:3921-31. [Crossref] [PubMed]
- Guo XG, Long JJ. Cytokeratin-19 fragment in the diagnosis of bladder carcinoma. Tumour Biol 2016;37:14329-30. [Crossref] [PubMed]
- Lei Q, Zhao L, Ye S, et al. Rapid and quantitative detection of urinary Cyfra21-1 using fluorescent nanosphere-based immunochromatographic test strip for diagnosis and prognostic monitoring of bladder cancer. Artif Cells Nanomed Biotechnol 2019;47:4266-72. [Crossref] [PubMed]
- Crew JP, O'Brien T, Bicknell R, et al. Urinary vascular endothelial growth factor and its correlation with bladder cancer recurrence rates. J Urol 1999;161:799-804. [Crossref] [PubMed]
- Jeon SH, Lee SJ, Chang SG. Clinical significance of urinary vascular endothelial growth factor in patients with superficial bladder tumors. Oncol Rep 2001;8:1265-7. [Crossref] [PubMed]
- Yang H, Wang Z, Guo Y, et al. Correlation and Significance of Urinary Soluble Fas and Vascular En-dothelial Growth Factor in Bladder Urothelial Cancer. Dis Markers 2015;2015:383509 [Crossref] [PubMed]
- Shimizu Y, Furuya H, Bryant Greenwood P, et al. A multiplex immunoassay for the non-invasive de-tection of bladder cancer. J Transl Med 2016;14:31. [Crossref] [PubMed]
- Gogalic S, Sauer U, Doppler S, et al. Validation of a protein panel for the noninvasive detection of re-current non-muscle invasive bladder cancer. Biomarkers 2017;22:674-81. [PubMed]
- Gogalic S, Sauer U, Doppler S, et al. Investigating Colorimetric Protein Array Assay Schemes for De-tection of Recurrence of Bladder Cancer. Biosensors (Basel) 2018;8:10. [Crossref] [PubMed]
- Furuya H, Pagano I, Chee K, et al. Comparison of Commercial ELISA Kits, a Prototype Multiplex Electrochemoluminescent Assay, and a Multiplex Bead-Based Immunoassay for Detecting a Urine-Based Bladder-Cancer-Associated Diagnostic Signature. Diagnostics (Basel) 2019;9:166. [Crossref] [PubMed]
- Batista R, Vinagre J, Prazeres H, et al. Validation of a Novel, Sensitive, and Specific Urine-Based Test for Recurrence Surveillance of Patients With Non-Muscle-Invasive Bladder Cancer in a Comprehensive Multicenter Study. Front Genet 2019;10:1237. [Crossref] [PubMed]
- Mian C, Pycha A, Wiener H, et al. Immunocyt: a new tool for detecting transitional cell cancer of the urinary tract. J Urol 1999;161:1486-9. [Crossref] [PubMed]
- Heicappell R, Müller M, Fimmers R, et al. Qualitative determination of urinary human complement factor H-related protein (hcfHrp) in patients with bladder cancer, healthy controls, and patients with benign urologic disease. Urol Int 2000;65:181-4. [Crossref] [PubMed]
- Smrkolj T, Mihelič M, Sedlar A, et al. Performance of nuclear matrix protein 22 urine marker and voided urine cytology in the detection of urinary bladder tumors. Clin Chem Lab Med 2011;49:311-6. [Crossref] [PubMed]
- O'Sullivan P, Sharples K, Dalphin M, et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. J Urol 2012;188:741-7. [Crossref] [PubMed]
- Pichler R, Fritz J, Tulchiner G, et al. Increased accuracy of a novel mRNA-based urine test for bladder cancer surveillance. BJU Int 2018;121:29-37. [Crossref] [PubMed]
- Sarosdy MF, Schellhammer P, Bokinsky G, et al. Clinical evaluation of a multi-target fluorescent in situ hybridization assay for detection of bladder cancer. J Urol 2002;168:1950-4. [Crossref] [PubMed]
- Liang Q, Zhang G, Li W, et al. Comparison of the diagnostic performance of fluorescence in situ hy-bridization (FISH), nuclear matrix protein 22 (NMP22), and their combination model in bladder car-cinoma detection: a systematic review and meta-analysis. Onco Targets Ther 2018;12:349-58. [Crossref] [PubMed]
- Kumar A, Kumar R, Gupta NP. Comparison of NMP22 BladderChek test and urine cytology for the detection of recurrent bladder cancer. Jpn J Clin Oncol 2006;36:172-5. [Crossref] [PubMed]
- Witjes JA, Morote J, Cornel EB, et al. Performance of the Bladder EpiCheck™ Methylation Test for Patients Under Surveillance for Non-muscle-invasive Bladder Cancer: Results of a Multicenter, Pro-spective, Blinded Clinical Trial. Eur Urol Oncol 2018;1:307-13. [Crossref] [PubMed]
- Larré S, Camparo P, Comperat E, et al. Diagnostic, staging, and grading of urothelial carcinomas from urine: performance of BCA-1, a mini-array comparative genomic hybridisation-based test. Eur Urol 2011;59:250-7. [Crossref] [PubMed]
- van Kessel KE, Beukers W, Lurkin I, et al. Validation of a DNA Methylation-Mutation Urine Assay to Select Patients with Hematuria for Cystoscopy. J Urol 2017;197:590-5. [Crossref] [PubMed]
- Ward DG, Bryan RT. Liquid biopsies for bladder cancer. Transl Androl Urol 2017;6:331-5. [Crossref] [PubMed]
- Fadl-Elmula I. Chromosomal changes in uroepithelial carcinomas. Cell Chromosome 2005;4:1. [Crossref] [PubMed]
- Panzeri E, Conconi D, Antolini L, et al. Chromosomal aberrations in bladder cancer: fresh versus for-malin fixed paraffin embedded tissue and targeted FISH versus wide microarray-based CGH analysis. PLoS One 2011;6:e24237 [Crossref] [PubMed]
- Matsuyama H, Ikemoto K, Eguchi S, et al. Copy number aberrations using multicolour fluorescence in situ hybridization (FISH) for prognostication in non-muscle-invasive bladder cancer (NIMBC). BJU Int 2014;113:662-7. [Crossref] [PubMed]
- Springer SU, Chen CH, Rodriguez Pena MDC, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife 2018;7:e32143 [Crossref] [PubMed]
- Ge G, Peng D, Guan B, et al. Urothelial Carcinoma Detection Based on Copy Number Profiles of Urinary Cell-Free DNA by Shallow Whole-Genome Sequencing. Clin Chem 2020;66:188-98. [Crossref] [PubMed]
- Bubendorf L, Grilli B, Sauter G, et al. Multiprobe FISH for enhanced detection of bladder cancer in voided urine specimens and bladder washings. Am J Clin Pathol 2001;116:79-86. [Crossref] [PubMed]
- Skacel M, Pettay JD, Tsiftsakis EK, et al. Validation of a multicolor interphase fluorescence in situ hybridization assay for detection of transitional cell carcinoma on fresh and archival thin-layer, liq-uid-based cytology slides. Anal Quant Cytol Histol 2001;23:381-7. [PubMed]
- Mian C, Lodde M, Comploj E, et al. Liquid-based cytology as a tool for the performance of uCyt+ and Urovysion Multicolour-FISH in the detection of urothelial carcinoma. Cytopathology 2003;14:338-42. [Crossref] [PubMed]
- Chuang KL, Chuang HC, Ng KF, et al. Urinary fluorescence in situ hybridization assay for detecting urothelial carcinoma in Taiwanese patients. BJU Int 2010;105:1413-6. [Crossref] [PubMed]
- Maffezzini M, Campodonico F, Capponi G, et al. Prognostic significance of fluorescent in situ hybridi-sation in the follow-up of non-muscle-invasive bladder cancer. Anticancer Res 2010;30:4761-5. [PubMed]
- Dimashkieh H, Wolff DJ, Smith TM, et al. Evaluation of urovysion and cytology for bladder cancer detection: a study of 1835 paired urine samples with clinical and histologic correlation. Cancer Cytopathol 2013;121:591-7. [Crossref] [PubMed]
- Cussenot O, Sighar K, Mohammed M, et al. Detection of specific chromosomal aberrations in urine using BCA-1 (oligo-CGH-array) enhances diagnostic sensitivity and predicts the aggressiveness of non-muscle-invasive bladder transitional cell carcinoma. World J Urol 2014;32:551-7. [Crossref] [PubMed]
- Léon P, Cancel Tassin G, Sighar K, et al. Prog Urol 2017;27:451-7. [Correlation of genetic and cytogenetic alterations in patho-logical aggressiveness urothelial carcinoma of the bladder: Performance of BCA-1, a mini-array com-parative genomic hybridisation-based test]. [Crossref] [PubMed]
- Chen CC, Shieh B, Jin YT, et al. Microarray profiling of gene expression patterns in bladder tumor cells treated with genistein. J Biomed Sci 2001;8:214-22. [Crossref] [PubMed]
- Mengual L, Burset M, Ars E, et al. DNA microarray expression profiling of bladder cancer allows identification of noninvasive diagnostic markers. J Urol 2009;182:741-8. [Crossref] [PubMed]
- Wikman FP, Lu ML, Thykjaer T, et al. Evaluation of the performance of a p53 sequencing microarray chip using 140 previously sequenced bladder tumor samples. Clin Chem 2000;46:1555-61. [Crossref] [PubMed]
- Mao L, Schoenberg MP, Scicchitano M, et al. Molecular detection of primary bladder cancer by mi-crosatellite analysis. Science 1996;271:659-62. [Crossref] [PubMed]
- Christensen M, Wolf H, Orntoft TF. Microsatellite alterations in urinary sediments from patients with cystitis and bladder cancer. Int J Cancer 2000;85:614-7. [Crossref] [PubMed]
- Togneri FS, Ward DG, Foster JM, et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur J Hum Genet 2016;24:1167-74. [Crossref] [PubMed]
- Ward DG, Bryan RT. Massively parallel sequencing of urinary DNA—the dawn of non-invasive bladder cancer detection and surveillance? Transl Cancer Res 2019;S204-7. [Crossref]
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 2015;15:25-41. [Crossref] [PubMed]
- Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer 2005;5:713-25. [Crossref] [PubMed]
- Jebar AH, Hurst CD, Tomlinson DC, et al. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene 2005;24:5218-25. [Crossref] [PubMed]
- Kompier LC, Lurkin I, van der Aa MN, et al. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One 2010;5:e13821 [Crossref] [PubMed]
- Serizawa RR, Ralfkiaer U, Steven K, et al. Integrated genetic and epigenetic analysis of bladder cancer reveals an additive diagnostic value of FGFR3 mutations and hypermethylation events. Int J Cancer 2011;129:78-87. [Crossref] [PubMed]
- Millholland JM, Li S, Fernandez CA, et al. Detection of low frequency FGFR3 mutations in the urine of bladder cancer patients using next-generation deep sequencing. Res Rep Urol 2012;4:33-40. [PubMed]
- Karnes RJ, Fernandez CA, Shuber AP. A noninvasive multianalyte urine-based diagnostic assay for urothelial cancer of the bladder in the evaluation of hematuria. Mayo Clin Proc 2012;87:835-42. [Crossref] [PubMed]
- Shore ND, Fernandez CA, Shuber AP. Noninvasive multianalyte diagnostic assay for monitoring bladder cancer recurrence. Res Rep Urol 2012;4:49-56. [PubMed]
- Fernandez CA, Millholland JM, Zwarthoff EC, et al. A noninvasive multi-analyte diagnostic assay: combining protein and DNA markers to stratify bladder cancer patients. Res Rep Urol 2012;4:17-26. [Crossref] [PubMed]
- Zuiverloon TC, Beukers W, van der Keur KA, et al. Combinations of urinary biomarkers for surveillance of patients with incident nonmuscle invasive bladder cancer: the European FP7 UROMOL project. J Urol 2013;189:1945-51. [Crossref] [PubMed]
- Critelli R, Fasanelli F, Oderda M, et al. Detection of multiple mutations in urinary exfoliated cells from male bladder cancer patients at diagnosis and during follow-up. Oncotarget 2016;7:67435-48. [Crossref] [PubMed]
- Ward DG, Baxter L, Gordon NS, et al. Multiplex PCR and Next Generation Sequencing for the Non-Invasive Detection of Bladder Cancer. PLoS One 2016;11:e0149756 [Crossref] [PubMed]
- Christensen E, Birkenkamp-Demtröder K, Nordentoft I, et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur Urol 2017;71:961-9. [Crossref] [PubMed]
- Ecke TH, Schlechte HH, Schulze G, et al. Four tumour markers for urinary bladder cancer--tissue polypeptide antigen (TPA), HER-2/neu (ERB B2), urokinase-type plasminogen activator receptor (uPAR) and TP53 mutation. Anticancer Res 2005;25:635-41. [PubMed]
- Xu X, Stower MJ, Reid IN, et al. Molecular screening of multifocal transitional cell carcinoma of the bladder using p53 mutations as biomarkers. Clin Cancer Res 1996;2:1795-800. [PubMed]
- Ecke TH, Lenk SV, Schlechte HH, et al. Tissue polypeptide antigen (TPA) in comparison with muta-tions of tumour suppressor gene P53 (TP53) in patients with bladder cancer. Anticancer Res 2003;23:957-62. [PubMed]
- Eissa S, Zohny SF, Zekri AR, et al. Diagnostic value of fibronectin and mutant p53 in the urine of patients with bladder cancer: impact on clinicopathological features and disease recurrence. Med Oncol 2010;27:1286-94. [Crossref] [PubMed]
- Scott SN, Ostrovnaya I, Lin CM, et al. Next-generation sequencing of urine specimens: A novel platform for genomic analysis in patients with non-muscle-invasive urothelial carcinoma treated with bacille Calmette-Guérin. Cancer Cytopathol 2017;125:416-26. [Crossref] [PubMed]
- Sailer V, Eng KW, Zhang T, et al. Integrative Molecular Analysis of Patients With Advanced and Met-astatic Cancer. JCO Precis Oncol 2019;3:PO.19.00047.
- Kinde I, Munari E, Faraj SF, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res 2013;73:7162-7. [Crossref] [PubMed]
- Isharwal S, Audenet F, Drill E, et al. Prognostic Value of TERT Alterations, Mutational and Copy Number Alterations Burden in Urothelial Carcinoma. Eur Urol Focus 2019;5:201-4. [Crossref] [PubMed]
- Stasik S, Salomo K, Heberling U, et al. Evaluation of TERT promoter mutations in urinary cell-free DNA and sediment DNA for detection of bladder cancer. Clin Biochem 2019;64:60-3. [Crossref] [PubMed]
- Ward DG, Gordon NS, Boucher RH, et al. Targeted deep sequencing of urothelial bladder cancers and associated urinary DNA: a 23-gene panel with utility for non-invasive diagnosis and risk stratification. BJU Int 2019;124:532-44. [Crossref] [PubMed]
- Allory Y, Beukers W, Sagrera A, et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: high frequency across stages, detection in urine, and lack of association with outcome. Eur Urol 2014;65:360-6. [Crossref] [PubMed]
- Eich ML, Rodriguez Pena MDC, Springer SU, et al. Incidence and distribution of UroSEEK gene panel in a multi-institutional cohort of bladder urothelial carcinoma. Mod Pathol 2019;32:1544-50. [Crossref] [PubMed]
- Patel KM, van der Vos KE, Smith CG, et al. Association Of Plasma And Urinary Mutant DNA With Clinical Outcomes In Muscle Invasive Bladder Cancer. Sci Rep 2017;7:5554. [Crossref] [PubMed]
- Birkenkamp-Demtröder K, Nordentoft I, Christensen E, et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur Urol 2016;70:75-82. [Crossref] [PubMed]
- Wild PJ, Fuchs T, Stoehr R, et al. Detection of urothelial bladder cancer cells in voided urine can be improved by a combination of cytology and standardized microsatellite analysis. Cancer Epidemiol Biomarkers Prev 2009;18:1798-806. [Crossref] [PubMed]
- Chan MW, Chan LW, Tang NL, et al. Hypermethylation of multiple genes in tumor tissues and voided urine in urinary bladder cancer patients. Clin Cancer Res 2002;8:464-70. [PubMed]
- Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst 2006;98:996-1004. [Crossref] [PubMed]
- Urakami S, Shiina H, Enokida H, et al. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res 2006;12:2109-16. [Crossref] [PubMed]
- Yates DR, Rehman I, Meuth M, et al. Methylational urinalysis: a prospective study of bladder cancer patients and age stratified benign controls. Oncogene 2006;25:1984-8. [Crossref] [PubMed]
- Yu J, Zhu T, Wang Z, et al. A novel set of DNA methylation markers in urine sediments for sensi-tive/specific detection of bladder cancer. Clin Cancer Res 2007;13:7296-304. [Crossref] [PubMed]
- Friedrich MG, Toma MI, Chun JK, et al. DNA methylation on urinalysis and as a prognostic marker in urothelial cancer of the bladder. Urologe A 2007;46:761-8. [Crossref] [PubMed]
- Sun J, Chen Z, Zhu T, et al. Hypermethylated SFRP1, but none of other nine genes “informative” for western countries, is valuable for bladder cancer detection in Mainland China. J Cancer Res Clin Oncol 2009;135:1717-27. [Crossref] [PubMed]
- Vinci S, Giannarini G, Selli C, et al. Quantitative methylation analysis of BCL2, hTERT, and DAPK promoters in urine sediment for the detection of non-muscle-invasive urothelial carcinoma of the bladder: a prospective, two-center validation study. Urol Oncol 2011;29:150-6. [Crossref] [PubMed]
- Costa VL, Henrique R, Danielsen SA, et al. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res 2010;16:5842-51. [Crossref] [PubMed]
- Renard I, Joniau S, van Cleynenbreugel B, et al. Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the non-invasive detection of primary bladder cancer in urine samples. Eur Urol 2010;58:96-104. [Crossref] [PubMed]
- Lin HH, Ke HL, Huang SP, et al. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of E-cadherin, p16, p14, RASSF1A genes in urine. Urol Oncol 2010;28:597-602. [Crossref] [PubMed]
- Chen PC, Tsai MH, Yip SK, et al. Distinct DNA methylation epigenotypes in bladder cancer from different Chinese sub-populations and its implication in cancer detection using voided urine. BMC Med Genomics 2011;4:45. [Crossref] [PubMed]
- Chung W, Bondaruk J, Jelinek J, et al. Detection of bladder cancer using novel DNA methylation bi-omarkers in urine sediments. Cancer Epidemiol Biomarkers Prev 2011;20:1483-91. [Crossref] [PubMed]
- Scher MB, Elbaum MB, Mogilevkin Y, et al. Detecting DNA methylation of the BCL2, CDKN2A and NID2 genes in urine using a nested methylation specific polymerase chain reaction assay to predict bladder cancer. J Urol 2012;188:2101-7. [Crossref] [PubMed]
- Reinert T, Borre M, Christiansen A, et al. Diagnosis of bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PLoS One 2012;7:e46297 [Crossref] [PubMed]
- Eissa S, Zohny SF, Shehata HH, et al. Urinary retinoic acid receptor-β2 gene promoter methylation and hyaluronidase activity as noninvasive tests for diagnosis of bladder cancer. Clin Biochem 2012;45:402-7. [Crossref] [PubMed]
- Costa VL, Henrique R, Danielsen SA, et al. TCF21 and PCDH17 methylation: An innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics 2011;6:1120-30. [Crossref] [PubMed]
- Reinert T, Modin C, Castano FM, et al. Comprehensive genome methylation analysis in bladder cancer: identification and validation of novel methylated genes and application of these as urinary tumor markers. Clin Cancer Res 2011;17:5582-92. [Crossref] [PubMed]
- Kandimalla R, Masius R, Beukers W, et al. A 3-plex methylation assay combined with the FGFR3 mu-tation assay sensitively detects recurrent bladder cancer in voided urine. Clin Cancer Res 2013;19:4760-9. [Crossref] [PubMed]
- Chihara Y, Kanai Y, Fujimoto H, et al. Diagnostic markers of urothelial cancer based on DNA meth-ylation analysis. BMC Cancer 2013;13:275. [Crossref] [PubMed]
- Yegin Z, Gunes S, Buyukalpelli R. Hypermethylation of TWIST1 and NID2 in tumor tissues and voided urine in urinary bladder cancer patients. DNA Cell Biol 2013;32:386-92. [Crossref] [PubMed]
- Maldonado L, Brait M, Michailidi C, et al. An epigenetic marker panel for recurrence risk prediction of low grade papillary urothelial cell carcinoma (LGPUCC) and its potential use for surveillance after transurethral resection using urine. Oncotarget 2014;5:5218-33. [Crossref] [PubMed]
- Hayashi M, Bernert H, Kagohara LT, et al. Epigenetic inactivation of VGF associated with Urothelial Cell Carcinoma and its potential as a non-invasive biomarker using urine. Oncotarget 2014;5:3350-61. [Crossref] [PubMed]
- Andersson E, Steven K, Guldberg P. Size-based enrichment of exfoliated tumor cells in urine increases the sensitivity for DNA-based detection of bladder cancer. PLoS One 2014;9:e94023 [Crossref] [PubMed]
- Su SF, de Castro Abreu AL, Chihara Y, et al. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladder cancer recurrence. Clin Cancer Res 2014;20:1978-89. [Crossref] [PubMed]
- Abern MR, Owusu R, Inman BA. Clinical performance and utility of a DNA methylation urine test for bladder cancer. Urol Oncol 2014;32:51.e21-6. [Crossref] [PubMed]
- Yeh CM, Chen PC, Hsieh HY, et al. Methylomics analysis identifies ZNF671 as an epigenetically re-pressed novel tumor suppressor and a potential non-invasive biomarker for the detection of urothelial carcinoma. Oncotarget 2015;6:29555-72. [Crossref] [PubMed]
- Fantony JJ, Abern MR, Gopalakrishna A, et al. Multi-institutional external validation of urinary TWIST1 and NID2 methylation as a diagnostic test for bladder cancer. Urol Oncol 2015;33:387.e1-6. [Crossref] [PubMed]
- Roperch JP, Grandchamp B, Desgrandchamps F, et al. Promoter hypermethylation of HS3ST2, SEP-TIN9 and SLIT2 combined with FGFR3 mutations as a sensitive/specific urinary assay for diagnosis and surveillance in patients with low or high-risk non-muscle-invasive bladder cancer. BMC Cancer 2016;16:704. [Crossref] [PubMed]
- Wang K, Tian Y, Xu H. Improved Noninvasive Bladder Cancer Diagnosis using Urine Sediments and Novel DNA Methylation Biomarker Panels. Clin Lab 2016;62:327-36. [Crossref] [PubMed]
- Wang Y, Yu Y, Ye R, et al. An epigenetic biomarker combination of PCDH17 and POU4F2 detects bladder cancer accurately by methylation analyses of urine sediment DNA in Han Chinese. Oncotarget 2016;7:2754-64. [Crossref] [PubMed]
- Feber A, Dhami P, Dong L, et al. UroMark-a urinary biomarker assay for the detection of bladder cancer. Clin Epigenetics 2017;9:8. [Crossref] [PubMed]
- Pietrusiński M, Kȩpczyński Ƚ, Jȩdrzejczyk A, et al. Detection of bladder cancer in urine sediments by a hypermethylation panel of selected tumor suppressor genes. Cancer Biomark 2017;18:47-59. [Crossref] [PubMed]
- van der Heijden AG, Mengual L, Ingelmo-Torres M, et al. Urine cell-based DNA methylation classifier for monitoring bladder cancer. Clin Epigenetics 2018;10:71. [Crossref] [PubMed]
- Guo RQ, Xiong GY, Yang KW, et al. Detection of urothelial carcinoma, upper tract urothelial carci-noma, bladder carcinoma, and urothelial carcinoma with gross hematuria using selected urine-DNA methylation biomarkers: A prospective, single-center study. Urol Oncol 2018;36:342.e15-e23. [Crossref] [PubMed]
- Mijnes J, Veeck J, Gaisa NT, et al. Promoter methylation of DNA damage repair (DDR) genes in human tumor entities: RBBP8/CtIP is almost exclusively methylated in bladder cancer. Clin Epigenetics 2018;10:15. [Crossref] [PubMed]
- Steinbach D, Kaufmann M, Hippe J, et al. High Detection Rate for Non-Muscle-Invasive Bladder Cancer Using an Approved DNA Methylation Signature Test. Clin Genitourin Cancer 2020;18:210-21. [Crossref] [PubMed]
- Zhang N, Chen S, Wu L, et al. Identification of Cancer-Specific Methylation of Gene Combination for the Diagnosis of Bladder Cancer. J Cancer 2019;10:6761-6. [Crossref] [PubMed]
- Trenti E, D'Elia C, Mian C, et al. Diagnostic predictive value of the Bladder EpiCheck test in the fol-low-up of patients with non-muscle-invasive bladder cancer. Cancer Cytopathol 2019;127:465-9. [Crossref] [PubMed]
- Bosschieter J, Nieuwenhuijzen JA, Hentschel A, et al. A two-gene methylation signature for the diagnosis of bladder cancer in urine. Epigenomics 2019;11:337-47. [Crossref] [PubMed]
- Wu Y, Jiang G, Zhang N, et al. HOXA9, PCDH17, POU4F2, and ONECUT2 as a Urinary Biomarker Combination for the Detection of Bladder Cancer in Chinese Patients with Hematuria. Eur Urol Focus 2020;6:284-91. [Crossref] [PubMed]
- Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011;8:467-77. [Crossref] [PubMed]
- Fuessel S, Lohse-Fischer A, Vu Van D, et al. Quantification of MicroRNAs in Urine-Derived Specimens. Methods Mol Biol 2018;1655:201-26. [Crossref] [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [Crossref] [PubMed]
- Shimizu T, Suzuki H, Nojima M, et al. Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur Urol 2013;63:1091-100. [Crossref] [PubMed]
- Mengual L, Lozano JJ, Ingelmo-Torres M, et al. Using microRNA profiling in urine samples to develop a non-invasive test for bladder cancer. Int J Cancer 2013;133:2631-41. [PubMed]
- Zhang DZ, Lau KM, Chan ES, et al. Cell-free urinary microRNA-99a and microRNA-125b are diag-nostic markers for the non-invasive screening of bladder cancer. PLoS One 2014;9:e100793 [Crossref] [PubMed]
- Zhou X, Zhang X, Yang Y, et al. Urinary cell-free microRNA-106b as a novel biomarker for detection of bladder cancer. Med Oncol 2014;31:197. [Crossref] [PubMed]
- Eissa S, Habib H, Ali E, et al. Evaluation of urinary miRNA-96 as a potential biomarker for bladder cancer diagnosis. Med Oncol 2015;32:413. [Crossref] [PubMed]
- Wei S, Bing Z, Yao Y, et al. Higher Expression of miR-182 in Cytology Specimens of High-Grade Urothelial Cell Carcinoma: A Potential Diagnostic Marker. Acta Cytol 2015;59:109-12. [Crossref] [PubMed]
- Liu W, Qi L, Lv H, et al. MiRNA-141 and miRNA-200b are closely related to invasive ability and con-sidered as decision-making biomarkers for the extent of PLND during cystectomy. BMC Cancer 2015;15:92. [Crossref] [PubMed]
- Liu Q, Shin Y, Kee JS, et al. Mach-Zehnder interferometer (MZI) point-of-care system for rapid mul-tiplexed detection of microRNAs in human urine specimens. Biosens Bioelectron 2015;71:365-72. [Crossref] [PubMed]
- Zhang X, Zhang Y, Liu X, et al. Direct quantitative detection for cell-free miR-155 in urine: a potential role in diagnosis and prognosis for non-muscle invasive bladder cancer. Oncotarget 2016;7:3255-66. [Crossref] [PubMed]
- Sapre N, Macintyre G, Clarkson M, et al. A urinary microRNA signature can predict the presence of bladder urothelial carcinoma in patients undergoing surveillance. Br J Cancer 2016;114:454-62. [Crossref] [PubMed]
- Pospisilova S, Pazourkova E, Horinek A, et al. MicroRNAs in urine supernatant as potential non-invasive markers for bladder cancer detection. Neoplasma 2016;63:799-808. [Crossref] [PubMed]
- Urquidi V, Netherton M, Gomes-Giacoia E, et al. A microRNA biomarker panel for the non-invasive detection of bladder cancer. Oncotarget 2016;7:86290-9. [Crossref] [PubMed]
- Ingelmo-Torres M, Lozano JJ, Izquierdo L, et al. Urinary cell microRNA-based prognostic classifier for non-muscle invasive bladder cancer. Oncotarget 2017;8:18238-47. [Crossref] [PubMed]
- Du L, Jiang X, Duan W, et al. Cell-free microRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Oncotarget 2017;8:40832-42. [Crossref] [PubMed]
- Shindo T, Shimizu T, Nojima M, et al. Evaluation of Urinary DNA Methylation as a Marker for Re-current Bladder Cancer: A 2-Center Prospective Study. Urology 2018;113:71-8. [Crossref] [PubMed]
- Juracek J, Peltanova B, Dolezel J, et al. Genome-wide identification of urinary cell-free microRNAs for non-invasive detection of bladder cancer. J Cell Mol Med 2018;22:2033-8. [Crossref] [PubMed]
- Pardini B, Cordero F, Naccarati A, et al. microRNA profiles in urine by next-generation sequencing can stratify bladder cancer subtypes. Oncotarget 2018;9:20658-69. [Crossref] [PubMed]
- Piao XM, Jeong P, Kim YH, et al. Urinary cell-free microRNA biomarker could discriminate bladder cancer from benign hematuria. Int J Cancer 2019;144:380-8. [Crossref] [PubMed]
- Hofbauer SL, de Martino M, Lucca I, et al. A urinary microRNA (miR) signature for diagnosis of bladder cancer. Urol Oncol 2018;36:531.e1-e8. [Crossref] [PubMed]
- Nekoohesh L, Modarressi MH, Mowla SJ, et al. Expression profile of miRNAs in urine samples of bladder cancer patients. Biomark Med 2018;12:1311-21. [Crossref] [PubMed]
- Baumgart S, Meschkat P, Edelmann P, et al. MicroRNAs in tumor samples and urinary extracellular vesicles as a putative diagnostic tool for muscle-invasive bladder cancer. J Cancer Res Clin Oncol 2019;145:2725-36. [Crossref] [PubMed]
- Jiang F, Li C, Han J, et al. Diagnostic Value of Combination of MicroRNA-192 in Urinary Sediment and B-Ultrasound for Bladder Cancer. Technol Cancer Res Treat 2020;19:1533033819894573 [Crossref] [PubMed]
- Srivastava AK, Singh PK, Rath SK, et al. Appraisal of diagnostic ability of UCA1 as a biomarker of carcinoma of the urinary bladder. Tumour Biol 2014;35:11435-42. [Crossref] [PubMed]
- Milowich D, Le Mercier M, De Neve N, et al. Diagnostic value of the UCA1 test for bladder cancer detection: a clinical study. Springerplus 2015;4:349. [Crossref] [PubMed]
- Eissa S, Matboli M, Essawy NO, et al. Rapid detection of urinary long non-coding RNA urothelial carcinoma associated one using a PCR-free nanoparticle-based assay. Biomarkers 2015;20:212-7. [Crossref] [PubMed]
- Gielchinsky I, Gilon M, Abu-Lail R, et al. H19 non-coding RNA in urine cells detects urothelial carci-noma: a pilot study. Biomarkers 2017;22:661-6. [PubMed]
- Du L, Duan W, Jiang X, et al. Cell-free lncRNA expression signatures in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med 2018;22:2838-45. [Crossref] [PubMed]
- Song Z, Zhang Q, Zhu J, et al. Identification of urinary hsa_circ _0137439 as potential biomarker and tumor regulator of bladder cancer. Neoplasma 2020;67:137-46. [Crossref] [PubMed]
- Yu X, Wang R, Han C, et al. A Panel of Urinary Long Non-coding RNAs Differentiate Bladder Cancer from Urocystitis. J Cancer 2020;11:781-7. [Crossref] [PubMed]
- Wang XS, Zhang Z, Wang HC, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res 2006;12:4851-8. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics 2019;20:675-91. [Crossref] [PubMed]
- Chen X, Chen RX, Wei WS, et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res 2018;24:6319-30. [Crossref] [PubMed]
- Okholm TLH, Nielsen MM, Hamilton MP, et al. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom Med 2017;2:36. [Crossref] [PubMed]
- Liu L, Wu SQ, Zhu X, et al. Analysis of ceRNA network identifies prognostic circRNA biomarkers in bladder cancer. Neoplasma 2019;66:736-45. [Crossref] [PubMed]
- Goel A, Ward DG, Gordon NS, et al. Back-Splicing Transcript Isoforms (Circular RNAs) Affect Bio-logically Relevant Pathways and Offer an Additional Layer of Information to Stratify NMIBC Patients. Front Oncol 2020;10:812. [Crossref] [PubMed]
- Mostafid H, Babjuk M, Bochner B, et al. Transurethral Resection of Bladder Tumour: The Neglected Procedure in the Technology Race in Bladder Cancer. Eur Urol 2020;77:669-70. [Crossref] [PubMed]
- Wroblewski LE, Peek RM Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev 2010;23:713-39. [Crossref] [PubMed]
- Wu P, Zhang G, Zhao J, et al. Profiling the Urinary Microbiota in Male Patients With Bladder Cancer in China. Front Cell Infect Microbiol 2018;8:167. [Crossref] [PubMed]
- Mai G, Chen L, Li R, et al. Common Core Bacterial Biomarkers of Bladder Cancer Based on Multiple Datasets. Biomed Res Int 2019;2019:4824909 [Crossref] [PubMed]
- Liu F, Liu A, Lu X, et al. Dysbiosis signatures of the microbial profile in tissue from bladder cancer. Cancer Med 2019;8:6904-14. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Yoshida K, Tsuda M, Matsumoto R, et al. Exosomes containing ErbB2/CRK induce vascular growth in premetastatic niches and promote metastasis of bladder cancer. Cancer Sci 2019;110:2119-32. [Crossref] [PubMed]
- Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics 2010;9:1324-38. [Crossref] [PubMed]
- Thane KE, Davis AM, Hoffman AM. Improved methods for fluorescent labeling and detection of single extracellular vesicles using nanoparticle tracking analysis. Scientific Reports 2019;9:12295. [Crossref] [PubMed]
- Liang LG, Kong MQ, Zhou S, et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci Rep 2017;7:46224. [Crossref] [PubMed]



