
Epigenetics in prostate cancer: clinical implications
Introduction
Prostate cancer is the most common cancer in men in the United States, and it is estimated that over 34,000 men will die from prostate cancer in 2021 (1). There is therefore a need to better understand this malignancy and to develop new effective treatment strategies. Localized prostate cancer is clinically heterogeneous. Some men have indolent disease, and their prostate cancer is not likely to progress or cause death during their lifetime. Others have more aggressive disease that progresses to metastatic and lethal disease. In addition to clinical heterogeneity, there is molecular heterogeneity across patients with newly diagnosed prostate cancer, characterized by distinct genomic (e.g., TMPRSS2-ERG, SPOP, BRCA2) and transcriptomic differences (2-4).
For patients that develop metastatic disease, androgen deprivation therapy (ADT) represents the backbone of therapy, typically given in combination with docetaxel chemotherapy or potent androgen receptor (AR)-targeted drugs (5,6). For metastatic castration resistant prostate cancer (mCRPC), there are several approved drugs with varied mechanisms of action (7). Molecular biomarkers in advanced prostate cancer have largely focused on identifying genomic alterations that predict response to systemic therapies or identifying mechanisms of resistance. Most treatment resistant prostate cancers are still driven by AR signaling, but non-AR driven prostate cancer is an emerging subtype of therapy resistance.
In addition to genomic alterations, epigenetics plays a key role in prostate cancer initiation, progression, and treatment resistance. Epigenetics is the study of alterations, such as DNA methylation and histone modifications, which alter DNA accessibility and chromatin structure, thus influencing patterns of gene expression, but without altering the genetic code itself. Targeting epigenetic pathways is emerging therapeutic strategy in prostate cancer. Moreover, unique epigenetic patterns in cancer cells, such as specific DNA hypo- or hypermethylated sites or expression of key regulators, offer potential as diagnostic or predictive biomarkers (Figure 1). Here we review recent advances in the field and discuss the potential clinical implications of epigenetics across the prostate cancer continuum.
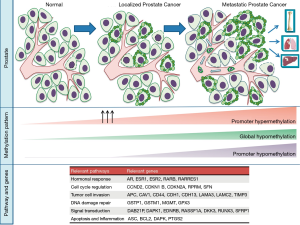
Role of epigenetics in localized prostate cancer
DNA methylation
Findings suggesting that DNA methylation contributes to prostate cancer development were reported in the early 1990s. In particular, cytidine methylation of regulatory sequences near the glutathione-S-transferase (GSTP1) gene was associated with prostate tumorigenesis (8). The GSTP1 gene encodes an enzyme involved in the protection of DNA from oxidants and carcinogens. In prostate cancer, GSTP1 is methylated at its promoter region which associates with loss of GSTP1 expression in tumor cells. Hypermethylation of the GSTP1 promotor occurs in approximately 75% of pre-invasive high-grade prostatic intraepithelial neoplasms, in more than 90% of prostate tumors, and persists through all stages of prostate cancer progression (9), suggesting that GSTP1 methylation is an early event in malignant transformation. In patients with localized disease, preoperative circulating methylated GSTP1 in plasma was shown to predict PSA recurrence and tumor aggressiveness (e.g., Gleason grade and tumor stage) (10-12).
In mammals, methylation mostly involves CpG dinucleotide regions of the DNA, and only about 1% of human genome has this dinucleotide due to spontaneous deamination of methylated CpG to TpG. However, there are CpG islands, which are short stretches of DNA enriched with CpG sequences. CpG islands occur predominantly at gene promotor regions near transcriptional start sites and may become hypermethylated or hypomethylated when cells become malignant to regulate downstream gene expression.
DNA methylation is regulated by DNA methyltransferases (DNMTs), including DNMT1, DNMT3A and DNMT3B, a family of enzymes involved in post-replicative methylation of newly synthesized DNA strands (8). In prostate cancer, DNMT1 and DNMT3B collaborate to silence specific genes across the genome and contribute to a malignant cellular phenotype (13,14).
DNA methyltransferase enzymes catalyze the addition of methyl groups to cytosine residues in DNA to form 5-methyl cytosine (5mC), which can be deleted by the TET family of DNA demethylases (15). TET family proteins favor locus-specific reversal of DNA methylation in normal cells, leading to the regulation of DNA methylation patterns and gene expression (16). In several solid tumors. including prostate cancer, TET-mediated 5hmC and expression of tumor-suppressive TET family proteins can be reduced (16). Downregulation of TET2 has been implicated in the regulation of AR signaling and prostate cancer development (17,18). Decreased expression of both TET2 and 5hmC associated with prostate cancer progression have been nominated as potential prognostic biomarkers (19,20).
Concurrent global hypomethylation and promoter hypermethylation has been suggested to play an important role in cellular transformation (21,22). In addition to GSTP1, hypermethylation of the promoter regions of other genes have been identified as early events in prostate cancer, including adenomatous polyposis coli (APC), Ras-associated domain family 1A (RASSF1), O6-methylguanine DNA methyltransferase (MGMT), cyclin-dependent kinase inhibitor 2A (CDKN2A), death-associated protein kinase (DAPK), and tissue inhibitors of metalloproteinases (TIMPS), and others (23-26). These genes are involved in diverse cellular pathways including invasion, metastasis, cell cycle, apoptosis, DNA repair, and hormonal response. The Cancer Genome Atlas (TCGA) study of 333 localized prostate tumors identified distinct methylation patterns underlying genomic subtypes (e.g., ERG-fusion positive, SPOP or FOXA1 mutated, and IDH1-mutant cancers) (4) suggesting an interplay between genomics and DNA methylation.
ConfirmMDx is a tissue-based DNA methylation assay developed for men with suspected prostate cancer to distinguish true negative prostate biopsies from occult prostate cancer. This epigenetic test evaluates methylation of three genes: GSTP1, APC, and RASSF2 (27). ConfirmMDx has been supported by National Comprehensive Cancer Network (NCCN) guidelines for men with at least one prior negative biopsy. In the Methylation Analysis to Locate Occult Cancer (MATLOC) trial, ConfirmMDx had a significant prognostic impact with an odds ratio of 3.17 (28). In the Detection of Cancer Using Methylated Events in Negative Tissue (DOCUMENT) study, the assay was independently associated with prostate cancer detection on repeat biopsy with an 88% negative predictive value (29). In the Prostate Assay Specific Clinical Utility at Launch (PASCUAL) study, the ConfirmMDx assay impacted repeat prostate biopsy decision-making in US urologic practices (30). Therefore, this assay is sometimes applied clinically in men with suspected prostate cancer with prostate biopsy demonstrating a cancer-free histopathology.
Histone modifications and other epigenetic alterations
In addition to DNA methylation, other epigenetic modifications influence gene expression in prostate cancer through chromatin acetylation and/or specific histone modifications (31). The process of histone acetylation at lysine residues is under the control of histone acetyl transferases (HATs) and histone deacetylases (HDACs) (32,33). In prostate cancer, transcription of AR target genes is regulated by the assembly of a complex of transcription factors. AR agonists promote the recruitment of the AR and coactivators that have histone acetyltransferase activity to promoters of AR target genes, leading to histone acetylation and active transcription (34). Conversely, AR bound to antagonists, such as bicalutamide, activates corepressors, such as NCoR or SMRT, that are joined to HDACs to suppress gene expression (35). Consistent with this evidence, HDAC inhibitors are capable of reversing transcriptional repression induced by nuclear receptors.
Histone lysine demethylases (KDMs) are a class of epigenetic enzymes that can remove both repressive and activating histone marks. KDMs are currently grouped into 7 major classes, each one targeting a specific methylation site. KDM expression has been found to be deregulated in prostate cancer playing a role as either tumor suppressors or oncogenes, depending on their gene regulatory function. Among KDMs, the lysine-specific demethylase 1A and 4C (KDM1A and KDM4C) associated with prostate cancer progression and androgen-dependent proliferation. Interaction of KDM4C and KDM1A with the AR can also regulate AR function through demethylation of histone H3 lysine 9 (36).
Through combined RNA-sequencing and ChIPseq, Stelloo et al. (37) identified three molecular subtypes of prostate cancer based on AR binding and histone modifications. Two of these subtypes were under TMPRSS2-ERG control and the third subtype was characterized by low chromatin binding and low AR activity and overactivity of FGF and WNT signaling. In a recent study by Pomerantz et al. (38), integration of the epigenome with genomic and transcriptome data discovered that primary prostate tumors can be distinguished from mCRPC based on transcription factor binding and metastasis specific histone acetylation (H3K27ac) patterns at enhancers including AR, FOXA1, and HOXB13. Interestingly, epigenomic programs that occur during prostate development were reactivated during metastatic progression.
Role of epigenetics in advanced prostate cancer
Castration-resistant prostate cancer represents a disease state that develops after tumors progress on initial androgen deprivation therapy. Despite significant advances in therapy for men with mCRPC, the median survival of mCRPC is approximately three years (39). Research in mCRPC has focused on developing new effective systemic therapies and in identifying mechanisms of drug resistance. While genomic and transcriptomic alterations have been largely explored, data on the epigenetic landscape of mCRPC is still accumulating.
DNA methylation
Variations in DNA methylation, either globally or locus specifically, are recurrent events in mCRPC (Figure 2). Promoter hypermethylation can silence not only tumor-suppressor genes, but also the androgen receptor (AR) and estrogen receptor (ESR1), cell adhesion genes (e.g., CD44, CDH1), cell-cycle genes (e.g., CCND2, CDKN1B, SFN), and apoptotic genes (e.g., ASC, BCL2, DAPK, PTGS2) (40). While global hypomethylation is frequent in primary tumors, it is even more evident in metastatic prostate cancers (41). Promoter hypomethylation can lead to activation of proto-oncogenes, such as PLAU (urokinase-type plasminogen activator) (42,43).
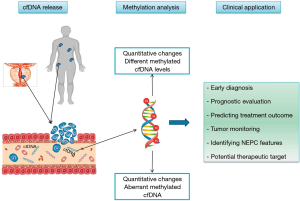
Whole genome bisulfite sequencing (WGBS) has recently been utilized to study methylated regions across the prostate cancer genome in mCRPC. While most prior studies in prostate cancer had evaluated a small percentage of the genome, predominantly analysing CpG islands at promoter regions, many important regulatory regions are outside of these areas. In a study by Zhao et al. (44), WGBS of mCRPC biopsies was performed with matched deep whole-genome sequencing (WGS) and RNA-sequencing (RNA-seq) in 100 patients. They observed that 22% of mCRPC tumors harbor distinct epigenomic features characterized by hypermethylation and enriched with somatic mutations involving TET2, DNMT3B, IDH1 and BRAF. In addition, differentially methylated region (DMR) analysis comparing benign prostate tissues versus primary tumors versus mCRPC identified cancer-associated hypo-methylation of specific regions driving over-expression of oncogenic drivers in mCRPC. In this study, localized prostate tumors were predominantly less methylated compared with benign prostate, and mCRPC samples were predominantly less methylated than primary tumors, with an overlap of 55% between the DMRs from localized versus metastatic disease. In general, the regions with the most differential hypomethylation in mCRPC were characterized by a higher somatic mutation rate, suggesting that certain regions of the genome are more commonly somatically altered by both mutation and methylation.
Histone modifications and other epigenetic alterations
Various other types of epigenetic changes have been described in mCRPC, including covalent modifications at specific amino acids on histones, including acetylation, methylation, phosphorylation, ADP-ribosylation, ubiquitination. These alterations mainly involve the N-terminal tails of histones. The added moieties contribute to the properties of the histone, loosening or tightening the condensed nucleosome structure. Modifications of histones can affect DNA-associated processes, including packaging, transcription regulation, replication, recombination, and repair. Studies also support their role in the progression of prostate cancer (45). In a study looking at global histone methylation patterns across the prostate cancer continuum, di- and trimethylation of H3K9 and acetylation of H3 and H4 were found to be lower, and mono-, di-, tri-methylation of H3K4 increased in CRPC (46).
In the recent study by Pomerantz et al. (38), the prostate epigenome was assessed across clinical states spanning from normal prostate epithelium to localized prostate cancer to metastatic castration resistant disease. This study identified epigenetically reprogrammed AR sites based on acetylation of the lysine 27 position on histone H3 (H3K27ac) that did not arise de novo. During the evolution from localized to mCRPC, there was reactivation of latent regulatory elements that were active during fetal prostate organogenesis. The integration of epigenetic and genetic reprogrammed regulatory loci in metastatic prostate tumors identified state-specific enhancers important for tumor progression. For example, among the different alterations explored in this study, gain of H3K27 acetylation coinciding with somatic DNA amplification identified metastasis-specific regulatory sites.
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of polycomb repressive complex 2 (PRC2) that acts to silence transcription through trimethylation of histone H3 lysine 27 (H3K27me3). EZH2 is overexpressed in various cancer types, including advanced prostate cancer (47). In addition to transcriptional silencing, EZH2 in CRPC can act as a coactivator of transcription factors such as the AR. AR activation requires phosphorylation of EZH2 and an intact methyltransferase domain. Consequently, EZH2 has been explored as a potential therapeutic target for mCRPC (48).
Lysine-specific demethylase 1 (LSD1), or KDM1A, is a histone demethylase that can regulate AR transcriptional activity through H3K4 demethylation (49). Moreover, LSD1 can demethylate the FOXA1 pioneer transcription factor, leading to FOXA1 activation which, in turn, mediates its recruitment to AR-dependent enhancers (50). LSD1 can also promote CRPC through regulation of CENPE, a centromere-binding protein and mitotic kinesin (51), as well as cooperation with the LSD1 binding protein, ZNF217 which promotes the expression of genes commonly not activated by androgens but involved in different pathways (cell-cycle, mitosis, and stemness) enriched in lethal prostate cancers (52). Other histone demethylases have also been implicated in prostate cancer progression. Lysine specific demethylase 5D (KDM5D) is a demethylase that influences AR signaling and response to docetaxel (53). Lysine specific demethylase 4B (KDM4B) promotes AR-V7 expression and resistance to AR-directed therapies (54). Lysine specific demethylase 3B (KDM3B) has been implicated in androgen-independent CRPC (55). Lysine specific demethylase eight (KDM8/JMJD5) promotes cellular proliferation and controls the activity of AR, HIF-1a, and EZH2 (56). These point to distinct regulation of key pathways by histone demethylases that may be leveraged therapeutically.
Bromodomain-containing (BRD) proteins are chromatin readers that recognize mono-acetylated histones, trigger chromatin remodeling, and initiate transcription. Dysregulation of bromodomains is common in cancer including metastatic prostate cancer (57). Moreover, BRD-containing proteins have diverse catalytic and scaffolding activities that function as transcription factors, transcriptional co-factors, methyltransferases, HATs, and helicases (57). Among the BRD family, BRD4 is a critical co-regulator of the AR (57,58), and BRD4 protein expression increases with castration resistance and associates with poor prognosis. BET inhibitors have demonstrated antitumor activity in CRPC models, attenuating AR signaling and reducing C-MYC expression (59,60). In addition, BET inhibitors have been shown to decrease AR-V7 expression by regulating splicing factors required for its generation (61). Taken together, these preclinical data have led to clinical trials of BET inhibitors as a therapeutic strategy to overcome persistent AR signaling in CRPC.
Among the proteins involved in histone modifications, p300 and CBP interact with transcription factors via their histone acetyltransferase domain (HAT) and bromodomain. In prostate cancer, p300 and CBP are highly homologous and frequently upregulated (62). Moreover, they have associated with disease progression and poor prognosis, representing potential therapeutic targets due to their function as AR coactivators and involvement in castration resistance (62). More recently, p300/CBP inhibition has been shown to inhibit secretion of exosomal PD-L1 by tumor cells (63), suggesting that the combination of HAT inhibitors with immunotherapy could play a synergic role in the treatment of prostate cancer.
Epigenetics and cell-free DNA
Tissue from localized prostate cancer for epigenetic profiling is relatively easy to access from radical prostatectomy or biopsy specimens, whereas collecting tissue biopsies from metastatic disease is more challenging. In recent years, epigenetic profiling assessing circulating cell free DNA (cfDNA) has been studied as an alternative to help overcome this challenge. In patients with metastatic prostate cancer, genomic profiles of circulating tumor DNA in plasma are closely matched with tumor metastasis tissues (64). A recent study characterized the mCRPC plasma methylome and genome concurrently (65) and found that the major contributor of methylation variance (principal component from the mCRPC plasma methylome analysis) in cfDNA was genomically determined circulating tumor DNA fraction (r =–0.96; P<10–8). Patient plasma samples with AR copy number gain had a significantly reduced AR- circulating tumor methylation signature ratio than AR copy number normal samples (P<0.001).
DNA methylation of cfDNA has also been investigated as a potential prognostic biomarker in mCRPC (12,65-71) (Table 1). Mahon et al. (68) showed that methylated GSTP1 became undetectable after two cycles in about 50% of patients with mCRPC treated with docetaxel chemotherapy. Undetectable methylated GSTP1 at baseline (20%) was correlated with longer overall survival (OS) (P<0.00001), and undetectable methylated GSTP1 after two cycles of chemotherapy was associated with a longer OS (P<0.00001) and time to PSA progression (P<0.00001). Peter et al. (70) focused on the dynamics of the cfDNA methylome of mCRPC patients during AR-directed therapies, showing that patients that maintained methylation during treatment had a longer time to clinical progression.

Full table
These studies point to the feasibility and potential clinical value in assessing plasma cfDNA methylation in patients with mCRPC. Future studies geared at molecular stratification, including in the context of epigenetic therapies, are warranted.
Role of epigenetics in neuroendocrine prostate cancer
Up to 15–20% of patients with mCRPC develop non-AR driven disease as a mechanism of treatment resistance, which has been associated in some cases with pathologic features of small cell neuroendocrine prostate cancer (NEPC). This form of prostate cancer is characterized by loss of AR signaling, expression of neuroendocrine lineage markers, and distinct changes in DNA methylation (72). The mammalian SWI/SNF chromatin remodeling complex is also dysregulated in NEPC (73). Integrative analyses of DNA methylation and transcriptome data points to epigenetic regulation of developmental, stem-like, and neuronal pathways in NEPC (73). In preclinical models, inactivation of RB1 and/or TP53 leads to upregulation of the DNA methyltransferase family member DNMT1, which is overexpressed in NEPC compared to prostate adenocarcinoma (74). Changes in DNA methylation patterns are particularly evident at gene loci of regulators of cell fate and neuronal development (75). Hypermethylation and reduced expression of the ETS family transcription factor SPDEF has been found in NEPC (76); SPDEF has been reported to be involved in suppression of tumor metastasis through inhibition of epithelial-mesenchymal transition (76). Methylation patterns of NEPC are also detectable in plasma cfDNA of patients (71), suggesting that methylome analysis of liquid biopsies might serve as a diagnostic or monitoring tool during treatment to identify patients developing NEPC (Figure 2).
Another epigenetic feature of NEPC is significant up-regulation of EZH2, even compared to mCRPC. In addition to its polycomb function, EZH2 can interact with N-myc to suppress AR signaling and drive NEPC progression (77). Genetic and pharmacological suppression of EZH2 inhibits NEPC growth and modulates plasticity including expression of AR signaling and neuroendocrine genes, potentially re-sensitizing tumors to AR-targeted drugs (72,74). Preclinical studies point to combination therapies with sub-lethal doses of the EZH2 inhibitor GSK343 and aurora kinase or epidermal growth factor receptor class of inhibitors against NEPC compared to CRPC (78). Following pre-clinical results in both CRPC and NEPC models, EZH2 inhibitors are in varied stages of clinical trials.
Epigenetic therapies
With an accumulating knowledge of the epigenetic alterations that drive prostate cancer progression and treatment resistance, epigenetic drugs are being developed for patients with advanced prostate cancer (Figure 3).
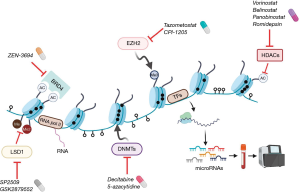
Targeting DNMT family members with drugs such as azacitidine and decitabine has been widely studied in myelodysplastic syndromes and other cancers (79). Alterations in DNA methylation can occur early in prostate cancer and aberrant DNMT expression occurs during prostate cancer progression (79). In a pre-clinical study, disulfiram, a DNMT inhibitor, suppressed cancer cell growth in vitro (DU145, CWR22R1, PC3 and C4-2B cell lines) and vivo (xenograft tumors of C4-2B) (80). Currently, drugs targeting DNMT in combination with chemotherapy, androgen receptor pathway inhibitor and immunotherapy, are in clinical development for prostate cancer. A phase I/II clinical trial of the DNMT inhibitor azacitidine in combination with docetaxel and prednisone for patients with mCRPC resulted in and 10 of 19 patients with PSA responses (>50% decline) and three patients with radiographic response (81). Understanding how DNA methylation profiles and/or identifying other alterations associate with responses to DNMT inhibition will be important considerations.
There are currently four FDA-approved HDAC inhibitors (vorinostat, belinostat, panobinostat and romidepsin) for the treatment of lymphoma and melanoma, and this class of drugs has also been investigated in prostate cancer (82). A phase II trial reported that combination treatment of panobinostat and bicalutamide prolonged radiographic progression free survival compared with bicalutamide alone in mCRPC patients without biomarker selection (83), suggesting that further studies may be warranted particularly in the context of novel AR-targeted drugs.
The polycomb group proteins EZH2 and embryonic ectoderm development (EED) (another member of the PRC2 complex) are currently being investigated as therapeutic targets. Tazometostat is an EZH2 inhibitor approved for follicular lymphoma (84). This agent is also being evaluated in a clinical trial for patients with mCRPC in combination with AR blockade (85). In addition, a selective EZH2 inhibitor (CPI-1205) is currently in clinical trials in combination with AR signaling inhibitors or immunotherapy (ipilimumab) (86). EED functions as a direct regulator of AR and AR downstream genes along with EZH2 in the context of AR-positive prostate cancer (87). MAK683 (an EED inhibitor) is being tested in a phase I/II clinical trial for advanced malignancies including relapsed prostate cancer whom no further effective standard treatment is available (88). While these trials are in biomarker-unselected populations, understanding their impact in AR-driven and non-AR driven CRPC populations may also be important.
Drugs targeting the bromodomain and extra-terminal (BET) family of chromatin readers, including the bromodomain containing protein (BRD) 2, BRD3, and BRD4 (58), are also in development. The clinical utility of a bromodomain extra-terminal (BET) inhibitor (ZEN-3694) was evaluated in combination with enzalutamide in 75 patients with mCRPC who had progressed after prior abiraterone acetate and/or enzalutamide (88). This phase Ib/IIa study revealed a median radiographic progression-free survival (rPFS) of 9.0 months and long term therapeutic effects without progression were observed in 17% (≥12 months) and 5% (≥24 months). In a subgroup analysis, patients with lower AR transcriptional activity in their baseline tumor biopsies had a longer rPFS (median rPFS: 10.4 vs. 4.3 months). Of note, a favorable rPFS was also seen in patients with aggressive variant clinical features (11.6 vs. 5.5 months).
CCS1477 is a potent, selective and orally active small molecule inhibitor of the bromodomain of p300/CBP. Its role has been explored for the treatment of CRPC through its downregulation of the AR, AR-splice variants and c-Myc expression, and in association with loss of function mutations in p300 or CBP by driving synthetic lethality (89,90). Early phase trials in mCRPC are ongoing.
A small-molecule LSD1 inhibitor SP2509 attenuated tumor growth in CRPC xenograft models (52). Another study showed that inhibition of LSD1 disrupts chromatin binding of FOXA1 which is a pioneer transcription factor for AR (50). Through regulation of AR binding and gene expression, LSD1 inhibition suppressed tumor growth and also showed a synergistic effect with enzalutamide in CRPC models (52). These findings provide support for LSD1 as a potential therapeutic target and the LSD1 inhibitor INCB059872 is in clinical development.
Notably, clinical trials to date have been for biomarker-unselected populations. Understanding the tumor specificity and downstream effects of these epigenetic therapies may be important considerations to improve patient selection and combination approaches in the future.
Conclusions
Epigenetic dysregulation, including changes in DNA methylation, histone modifications and nucleosome remodeling occurs at every phase of prostate cancer development and progression. These aberrations are responsible for silencing tumor-suppressor genes, activating oncogenic drivers, and driving therapy resistance. Thus, epigenetic modifications provide exciting potential as prostate cancer biomarkers and therapeutic targets. Promoter hypermethylation is widespread in prostate cancer, and restoration of a 'normal' epigenome through specific inhibitors of epigenetic enzymes has significant biologic and clinical implications. Epigenetic patterns correlate with clinical and pathologic predictors of prostate cancer phenotype and outcomes. Ongoing clinical studies evaluating emerging epigenetic therapies hold promise as potential treatment strategies for patients with advanced prostate cancer.
Acknowledgments
Funding: HB is supported by the Prostate Cancer Foundation, National Cancer Institute R37 CA241486-01A1, and Department of Defense PCRP W81XWH-17-1-0653.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Badrinath R Konety, Daniel W. Lin) for the series “Current and Future Topics on Prostate Cancer” published in Translational Andrology and Urology. The article has undergone external peer review.
Peer Review File: Available at http://dx.doi.org/10.21037/tau-20-1339
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1339). The series “Current and Future Topics on Prostate Cancer” was commissioned by the editorial office without any funding or sponsorship. VC reports advisory/consulting from Janssen and Bayer, and speaker honoraria or travel support from Astellas, Janssen-Cilag, and Sanofi-Aventis, during the conduct of the study. YY reports grants from Uehara Memorial Foundation Fellowship, outside the submitted work. SYK reports grants from Department of Defense, grants from Prostate Cancer Foundation Young Investigator Award, outside the submitted work. HB reports advisory/consulting from Janssen, Amgen, Astra Zeneca, Pfizer, Astellas, Sanofi Genzyme and research funding from Janssen, Abbvie Stemcentryx, Eli Lilly, Millenium, Astellas, during the conduct of the study. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Boutros PC, Fraser M, Harding NJ, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 2015;47:736-45. [Crossref] [PubMed]
- Robinson D, Van Allen EM, Wu YM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015;162:454. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011-25. [Crossref] [PubMed]
- Parker C, Castro E, Fizazi K, et al. ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2020;31:1119-34. [Crossref] [PubMed]
- VanderWeele DJ, Antonarakis ES, Carducci MA, et al. Metastatic Hormone-Sensitive Prostate Cancer: Clinical Decision Making in a Rapidly Evolving Landscape of Life-Prolonging Therapy. J Clin Oncol 2019;37:2961-7. [Crossref] [PubMed]
- Parker C, Castro E, Fizazi K, et al. Prostate cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2020;31:1119-34. [Crossref] [PubMed]
- Stein R, Gruebaum Y, Pollack Y, Razin A, Cedar H. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc Natl Acad Sci 1982;79:61-5. [Crossref] [PubMed]
- Bastian PJ, Yegnasubramanian S, Palapattu GS, et al. Molecular biomarker in prostate cancer: the role of CpG island hypermethylation. Eur Urol 2004;46:698-708. [Crossref] [PubMed]
- Delgado-Cruzata L, Hruby GW, Gonzalez K, et al. DNA methylation changes correlate with Gleason score and tumor stage in prostate cancer. DNA Cell Biol 2012;31:187-92. [Crossref] [PubMed]
- Rouprêt M, Hupertan V, Catto JW, et al. Promoter hypermethylation in circulating blood cells identifies prostate cancer progression Int J Cancer 2008;122:952-6. [Crossref] [PubMed]
- Mahon KL, Qu W, Devaney J, et al. Methylated Glutathione S-transferase 1 (mGSTP1) is a potential plasma free DNA epigenetic marker of prognosis and response to chemotherapy in castrate-resistant prostate cancer. Br J Cancer 2014;111:1802-9. [Crossref] [PubMed]
- Leu Y-W, Rahmatpanah F, Shi H, et al. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res 2003;63:6110-5. [PubMed]
- Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 2002;416:552-6. [Crossref] [PubMed]
- Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and 515 cancer. Genes Dev 2016;30:733-50. [Crossref] [PubMed]
- Ficz G, Branco MR, Seisenberger S, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 2011;473:398-402. [Crossref] [PubMed]
- Jin SG, Jiang Y, Qiu R, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360-5. [Crossref] [PubMed]
- Nickerson ML, Das S, Im KM, et al. TET2 binds the androgen receptor and loss is associated with prostate cancer. Oncogene 2017;36:2172-83. [Crossref] [PubMed]
- Takayama K, Misawa A, Suzuki T, et al. TET2 repression by androgen hormone regulates global hydroxymethylation status and prostate cancer progression. Nat Commun 2015;6:8219. [Crossref] [PubMed]
- Hsu CH, Peng KL, Kang ML, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep 2012;2:568-79. [Crossref] [PubMed]
- Storebjerg TM, Strand SH, Hoyer S, et al. Dysregulation and prognostic potential of 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) levels in prostate cancer. Clin Epigenetics 2018;10:105. [Crossref] [PubMed]
- Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med 2018;378:1323-34. [Crossref] [PubMed]
- Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016;17:284-99. [Crossref] [PubMed]
- Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin Cancer Res 2002;8:514-9. [PubMed]
- Yamanaka M, Watanabe M, Yamada Y, et al. Altered methylation of multiple genes in carcinogenesis of the prostate. Int J Cancer 2003;106:382-7. [Crossref] [PubMed]
- Padar A, Sathyanarayana UG, Suzuki M, et al. Inactivation of cyclin D2 gene in prostate cancers by aberrant promoter methylation. Clin Cancer Res 2003;9:4730-4. [PubMed]
- Van Neste L, Groskopf J, Grizzle WE, et al. Epigenetic risk score improves prostate cancer risk assessment. Prostate 2017;77:1259-64. [Crossref] [PubMed]
- Stewart GD, Van Neste L, Delvenne P, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: Results of the matloc study. J Urol 2013;189:1110-6. [Crossref] [PubMed]
- Partin AW, Van Neste L, Klein EA, et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 2014;192:1081-7. [Crossref] [PubMed]
- Yonover P, Steyaert S, Cohen JJ, et al. Clinical utility study of confirms mdx for prostate cancer in a community urology practice. J Clin Oncol 2019;37:94. [Crossref]
- Perry AS, Watson RWG, Lawler M, Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol 2010;7:668-80. [Crossref] [PubMed]
- Abbas A, Gupta S. The role of histone deacetylases in prostate cancer. Epigenetics 2008;3:300-9. [Crossref] [PubMed]
- Gryder BE, Akbashev MJ, Rood MK, et al. Selectively targeting prostate cancer with antiandrogen equipped histone deacetylase inhibitors. ACS Chem Biol 2013;8:2550-60. [Crossref] [PubMed]
- Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell 2002;9:601-10. [Crossref] [PubMed]
- Nagy L, Kao H-Y, Chakravarti D, et al. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 1997;89:373-80. [Crossref] [PubMed]
- Crea F, Sun L, Mai A, et al. The emerging role of histone lysine demethylases in prostate cancer. Mol Cancer 2012;11:52. [Crossref] [PubMed]
- Stelloo S, Nevedomskaya E, Kim Y, et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat Commun 2018;9:4900. [Crossref] [PubMed]
- Pomerantz MM, Qiu X, Zhu Y, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat Genet 2020;52:790-9. [Crossref] [PubMed]
- Davies A, Conteduca V, Zoubeidi A, et al. Biological Evolution of Castration-resistant Prostate Cancer. Eur Urol Focus 2019;5:147-54. [Crossref] [PubMed]
- Graça I, Pereira-Silva E, Henrique R, Packham G, Crabb SJ, Jerónimo C. Epigenetic modulators as therapeutic targets in prostate cancer. Clin Epigenetics 2016;8:98. [Crossref] [PubMed]
- Yegnasubramanian S, Haffner MC, Zhang Y, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res 2008;68:8954-67. [Crossref] [PubMed]
- Dong Z, Saliganan AD, Meng H, et al. Prostate cancer cell-derived urokinase-type plasminogen activator contributes to intraosseous tumor growth and bone turnover. Neoplasia 2008;10:439-49. [Crossref] [PubMed]
- Helenius MA, Savinainen KJ, Bova GS, et al. Amplification of the urokinase gene and the sensitivity of prostate cancer cells to urokinase inhibitors. BJU Int 2006;97:404-9. [Crossref] [PubMed]
- Zhao SG, Chen WS, Li H, et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet 2020;52:778-89. [Crossref] [PubMed]
- Nightingale KP, O'Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev 2006;16:125-36. [Crossref] [PubMed]
- Ellinger J, Kahl P, von der Gathen J, et al. Global levels of histone modifications predict prostate cancer recurrence. Prostate 2010;70:61-9. [Crossref] [PubMed]
- Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002;419:624-9. [Crossref] [PubMed]
- Xu K, Wu ZJ, Groner AC, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012;338:1465-9. [Crossref] [PubMed]
- Cai C, He HH, Chen S, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 2011;20:457-71. [Crossref] [PubMed]
- Gao S, Chen S, Han D, et al. Chromatin binding of FOXA1 is promoted by LSD1-mediated demethylation in prostate cancer. Nat Genet 2020;52:1011-7. [Crossref] [PubMed]
- Liang Y, Ahmed M, Guo H, et al. LSD1-Mediated Epigenetic Reprogramming Drives CENPE Expression and Prostate Cancer Progression. Cancer Res 2017;77:5479-90. [Crossref] [PubMed]
- Sehrawat A, Gao L, Wang Y, et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci U S A. 2018;115:E4179-88. [Crossref] [PubMed]
- Komura K, Jeong SH, Hinohara K, et al. Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc Natl Acad Sci U S A 2016;113:6259-64. [Crossref] [PubMed]
- Duan L, Chen Z, Lu J, et al. Histone lysine demethylase KDM4B regulates the alternative splicing of the androgen receptor in response to androgen deprivation. Nucleic Acids Res 2019;47:11623-36. [Crossref] [PubMed]
- Saraç H, Morova T, Pires E, et al. Systematic characterization of chromatin modifying enzymes identifies KDM3B as a critical regulator in castration resistant prostate cancer. Oncogene 2020;39:2187-201. [Crossref] [PubMed]
- Wang HJ, Pochampalli M, Wang LY, et al. KDM8/JMJD5 as a dual coactivator of AR and PKM2 integrates AR/EZH2 network and tumor metabolism in CRPC. Oncogene 2019;38:17-32. [Crossref] [PubMed]
- Urbanucci A, Mills IG. Bromodomain-containing proteins in prostate cancer. Mol Cell Endocrinol 2018;462:31-40. [Crossref] [PubMed]
- Fujisawa T, Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol 2017;18:246-62. [Crossref] [PubMed]
- Asangani IA, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014;510:278-82. [Crossref] [PubMed]
- Asangani IA, Wilder-Romans K, Dommeti VL, et al. BET bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists in the treatment of prostate cancer. Mol Cancer Res 2016;14:324-31. [Crossref] [PubMed]
- Welti J, Sharp A, Yuan W, et al. Targeting Bromodomain and Extra-Terminal (BET) Family Proteins in Castration-Resistant Prostate Cancer (CRPC). Clin Cancer Res 2018;24:3149-62. [Crossref] [PubMed]
- Lasko LM, Jakob CG, Edalji RP, et al. Discovery of a selective catalytic p300/CBP 578 inhibitor that targets lineage-specific tumours. Nature 2017;550:128-32. [Crossref] [PubMed]
- Liu J, He D, Cheng L, et al. p300/CBP inhibition enhances the efficacy of programmed death-ligand 1 blockade treatment in prostate cancer. Oncogene 2020;39:3939-51. [Crossref] [PubMed]
- Wyatt AW, Annala M, Aggarwal R, et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst 2017;109:djx118 [Crossref] [PubMed]
- Wu A, Cremaschi P, Wetterskog D, et al. Genome-wide plasma DNA methylation features of metastatic prostate cancer. J Clin Invest 2020;130:1991-2000. [Crossref] [PubMed]
- Bastian PJ, Palapattu GS, Lin X, et al. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin Cancer Res 2005;11:4037-43. [Crossref] [PubMed]
- Hendriks RJ, Dijkstra S, Smit FP, et al. Epigenetic markers in circulating cell-free DNA as prognostic markers for survival of castration-resistant prostate cancer patients. Prostate 2018;78:336-42. [Crossref] [PubMed]
- Mahon KL, Qu W, Lin HM, et al. Serum Free Methylated Glutathione S-transferase 1 DNA Levels, Survival, and Response to Docetaxel in Metastatic, Castration-resistant Prostate Cancer: Post Hoc Analyses of Data from a Phase 3 Trial. Eur Urol 2019;76:306-12. [Crossref] [PubMed]
- Gordevičius J, Krisciunas A, Groot DE, et al. Cell-Free DNA Modification Dynamics in Abiraterone Acetate-Treated Prostate Cancer Patients. Clin Cancer Res 2018;24:3317-24. [Crossref] [PubMed]
- Peter MR, Bilenky M, Isserlin R, et al. Dynamics of the cell-free DNA methylome of metastatic castration resistant prostate cancer during androgen-targeting treatment. Epigenomics 2020;12:1317-32. [Crossref] [PubMed]
- Beltran H, Romanel A, Conteduca V, et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest 2020;130:1653-68. [Crossref] [PubMed]
- Beltran H, Prandi D, Mosquera JM, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298-305. [Crossref] [PubMed]
- Cyrta J, Augspach A, De Filippo MR, et al. Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat Commun 2020;11:5549. [Crossref] [PubMed]
- Ku SY, Rosario S, Wang Y, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017;355:78-83. [Crossref] [PubMed]
- Stricker SH, Götz M. DNA-Methylation: Master or Slave of Neural Fate Decisions? Front Neurosci 2018;12:5. [Crossref] [PubMed]
- Chen WY, Tsai YC, Yeh HL, et al. Loss of SPDEF and gain of TGFBI activity after androgen deprivation therapy promote EMT and bone metastasis of prostate cancer. Sci Signal 2017;10:eaam6826 [Crossref] [PubMed]
- Dardenne E, Beltran H, Benelli M, et al. N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell 2016;30:563-77. [Crossref] [PubMed]
- Puca L, Bareja R, Prandi D, et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat Commun 2018;9:2404. [Crossref] [PubMed]
- Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol 2009;10:223-32. [Crossref] [PubMed]
- Lin J, Haffner MC, Zhang Y, et al. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate 2011;71:333-43. [Crossref] [PubMed]
- Singal R, Ramachandran K, Gordian E, et al. Phase I/II study of azacitidine, docetaxel, and prednisone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel-based therapy. Clin Genitourin Cancer 2015;13:22-31. [Crossref] [PubMed]
- Kaushik D, Vashistha V, Isharwal S, et al. Histone deacetylase inhibitors in castration-resistant prostate cancer: molecular mechanism of action and recent clinical trials. Ther Adv Urol 2015;7:388-95. [Crossref] [PubMed]
- Ferrari AC, Alumkal JJ, Stein MN, et al. Epigenetic Therapy with Panobinostat Combined with Bicalutamide Rechallenge in Castration-Resistant Prostate Cancer. Clin Cancer Res 2019;25:52-63. [Crossref] [PubMed]
- Italiano A, Soria JC, Toulmonde M, et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649-59. [Crossref] [PubMed]
- Ge R, Wang Z, Montironi R, et al. Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann Oncol 2020;31:470-9. [Crossref] [PubMed]
- Davies A, Zoubeidi A, Selth LA. The epigenetic and transcriptional landscape of neuroendocrine prostate cancer. Endocr Relat Cancer 2020;27:R35-50. [Crossref] [PubMed]
- Liu Q, Wang G, Li Q, et al. Polycomb group proteins EZH2 and EED directly regulate androgen receptor in advanced prostate cancer. Int J Cancer 2019;145:415-26. [Crossref] [PubMed]
- Aggarwal RR, Schweizer MT, Nanus DM, et al. A Phase 1b/2a Study of the Pan-BET Bromodomain Inhibitor ZEN-3694 in Combination with Enzalutamide in Patients with Metastatic Castration Resistant Prostate Cancer. Clin Cancer Res 2020;26:5338-47. [Crossref] [PubMed]
- Pegg N, Worthington J, Young B, et al. Novel small molecule inhibitors of p300/CBP down-regulate androgen receptor (AR) and c-Myc for the treatment of prostate cancer and beyond. Cancer Res 2018;78:3991.
- Welti J, Sharp A, Brooks N, et al. Targeting the p300/CBP Axis in Lethal Prostate Cancer. Cancer Discov 2021;11:1118-37. [Crossref] [PubMed]


