
The effects of histone crotonylation and bromodomain protein 4 on prostate cancer cell lines
Introduction
Prostate cancer (PCa) is one of the most common malignant tumors of the genitourinary system in middle-aged and elderly men (1,2). The incidence of PCa in China has increased in recent years, and it has become a serious threat to men’s health (3). At present, androgen deprivation therapy (ADT) is commonly used clinically to treat patients with recurrence and advanced metastasis, but after 12–18 months, the disease can progress to castration-resistant PCa (CRPC), which can eventually lead to death (4). There is no effective treatment for CRPC, and thus new approaches in PCa therapy are urgently needed.
Histone lysine crotonylation (Kcr) is a new type of modification found in 2011 that has a similar structure to histone lysine acetylation (Kac). Studies have shown that it has a certain role in the regulation of transcription, protein stability, metabolic pathways, and in the occurrence and development of PCa (5,6). Recent studies have shown that there is a partial overlap between histone crotonylation sites and histone acetylation, indicating the similar biological functions of the two modifications (7). Histone modification needs to be recognized by the corresponding effector protein to function (8). Bromodomain-containing protein 4 (BRD4) is a member of the bromodomain and extra-terminal domain (BET) family of proteins and is also an effector protein of histone acetylation. BRD4 can bind to the sites of histone acetylation and interact with the androgen receptor signaling pathway to exert its effects on tumor cells (3,9,10). Nevertheless, the relationship between BRD4 and crotonylation modification is still unclear. We therefore collected the PCa tissues from 72 patients and the adjacent tissues of 7 patients, and detect the level of histone crotonylation immunohistochemistry. Moreover, BRD4 inhibitors including I-BET762, I-BET726, and CPI-203, with I-BET762 used a positive control [its inhibitory effect on tumors has been reported in other studies (11)], were used for the first time to explore the effects of BRD4 inhibitors on histone crotonylation and the potential functions of crotonylation in the occurrence of PCa. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tau-21-53).
Methods
Sample source
Specimen source
The tissue samples were purchased from Xi’an Alina Biotechnology Co., Ltd. (Xi’an, China), and included PCa tissue from 72 patients and adjacent tissue from 7 patients. The clinical staging (TNM staging) of the samples was as follows: 4 cases in stage 1, 40 cases in stage 2, 12 cases in stage 3, and 15 cases in stage 4. Meanwhile, the distribution of pathological classification was as follows: 5 cases in grade 1, well-differentiated and low malignancy; 17 cases in grade 2, medium differentiation and medium malignancy; 41 cases in grade 3, poor differentiation and high malignancy. Some of the tumor samples lacked clinical data concerning staging or grade.
Cell lines
The cell lines used in this study were human PCa cell lines, including PC-3, LNCaP, and C42B cells, purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). LNCaP and C42B are androgen-sensitive, whereas PC-3 is non-androgen sensitive.
Main reagents
BET762, I-BET726, and CPI-203 were purchased from MedChemExpress (NJ, USA), cell counting Kit-8 (CCK-8) was purchased from Bimake (Houston, TX, USA), and Transwell Chambers were purchased from Corning (Corning, NY, USA). Anti-histone crotonylation (Kcr) antibody, anti-histone acetylation modification (Kac) antibody, and anti-Histone H3 Lys18 crotonylation (H3K18cr) antibody at the 18th amino acid position of histone H3 were purchased from PTM-Biolabs (Hangzhou, China). Anti-actin (β-actin) antibody was purchased from Sigma-Aldrich (St. Louis, MO, USA), anti-histone H3 antibody and BRD4 antibody were purchased from Abcam (Cambridge, UK), and anti-histone acetylase P300 antibody was purchased from AbSci (Vancouver, WA, USA). Anti-histone acetylase GCN5 antibody, anti-Cyclin D1 antibody, anti-HDAC1 antibody, anti-HDAC2 antibody, anti-HDAC3 antibody, anti-ACSS2 antibody, anti-AR antibody, and anti-PSA antibody were purchased from Santa Cruz Biotechnology (Dallas, TX, USA), and C-MYC was purchased from Proteintech.
Immunohistochemical test
After dewaxing, rehydration, antigen retrieval, removal of peroxidase, blocking, incubation of primary antibodies and secondary antibodies, DAB color development, counterstaining, dehydration, and transparency, the sample was mounted and observed under an optical microscope. The dilution of Kcr was 1:150 while that of H4ac was 1:1,000. To assess the degree of positive staining of tissue sections, dark brown was considered strong positive (3 points), brown was moderately positive (2 points), light yellow was weakly positive (1 point), while the blue nuclei staining or no antigen expression was negative (0 points). The percentage of cells with different degrees of staining was calculated by the following formula: score=∑(PI×I) = (percentage of weakly positive cells ×1) + (percentage of moderately positive cells ×2) + (percentage of strong positive cells ×3). In this formula, PI represents the percentage of positive cells of the total number of cells in the sample, and I represents the staining intensity (0–3 points). The score ranged from 0 to 300, with a lower score indicating a weaker positivity.
Experimental groups
The three PCa cell lines used IC50 as the treatment concentration of I-BET762, I-BET726 and CPI-203. For example, the IC50 of I-BET762 was used for the treatments of the three I-BET762 groups, with the IC50 of PC-3 being 85 nmol/L, the IC50 of LNCaP being 52 nmol/L, and the IC50 of C42B being 63 nmol/L.
Western blotting
Electrophoresis followed by membrane transfer, blocking, and primary antibody and secondary antibody incubation was performed, after which the Odyssey CLX dual infrared laser imaging system (LI-COR Biosciences, Lincoln, NE, USA) was used for membrane imaging. The primary antibody dilutions were the following: anti-Pan-Kcr antibody 1:1,000, anti-Pan-Kac antibody 1:1,000, anti-H3K18cr antibody 1:1,000, anti-P300 antibody 1:1,000, anti-GCN5 antibody 1:500, anti-H3 antibody 1:1,000, and anti-β-actin antibody 1:2,000.
Qualitative real time polymerase chain reaction (qRT-PCR)
The total RNA was extracted, and reverse transcribed to obtain complement DNA (cDNA), β-actin was then used as the internal control, and the mRNA expression was detected by SYBR Green kit (TaKaRa, China). The primer sequences are shown in Table 1.
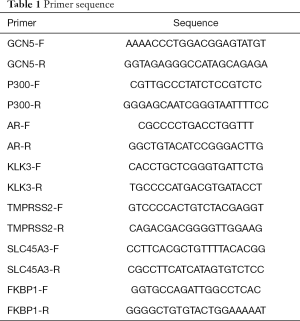
Full table
CCK-8 assay
A 0–5 d cell growth curve was made according to the kit instructions. Specifically, about 1×105 cells per well were added to the 96-well plate and cultured for the corresponding number of days. CCK-8 detection reagent and medium were mixed to a ratio of 1:10. The medium in the 96-well plate was then aspirated, and 100 µL of diluted reagent was added to each well. The 96-well plate was then incubated in a 37 °C incubator for 0.5–4 h until the color turned orange, the absorbance at 450 nm was then measured with a fluorescence counter, and a cell growth curve was made accordingly.
Cell cycles detected by flow cytometry
The treated cells were first digested with 0.25% trypsin, collected in a flow tube, resuspended in 70% ethanol, and fixed overnight at 4 °C in the dark. After fixation, RNaseA (final concentration 100 U/mL) was then added to the cell suspension, and incubated at 37 °C for 40 min in the dark, after which propidium iodide (PI) (final concentration 50 µg/mL) was added, and incubated at 37 °C for another 30 min in the dark before the flow cytometry was used for the detection of cell cycles.
Cell scratch test
About 5×105 cells per well were added to the 6-well plate and incubated overnight in a 37 °C incubator until the cells are confluent. A 1 mL pipette tip was used to make a scratch in the middle of the cells in the 6-well plate. The cells were then washed away with phosphate-buffered saline (PBS), and serum-free medium was added afterward. The pictures and samples were taken at 0, 12, and 24 h time points, and ImageJ software (US National Institutes of Health, Bethesda, MD, USA) was used to calculate the cell migration distance in the corresponding sampling point.
Transwell experiment
Cell suspension density was adjusted to [1–5]×105/mL with a serum-free medium (adjusted according to different cell migration and invasion capabilities). Next, 100–200 µL of these cell suspensions were added into the upper chamber of the Transwell in a 24-well plate and given different treatments, and 500 µL of medium containing fetal bovine serum was then added to the lower chamber. The cells were incubated in a 37 °C incubator for 12 to 48 h. After incubation, cells were then fixed with 4% paraformaldehyde, and the cells in the upper chamber were wiped off, stained with 0.1% crystal violet at room temperature, and decolorized with 33% acetic acid. The eluate was then used to measure the absorbance value at 570 nm with a microplate reader. The experiment was repeated 3 times.
Statistical analysis
SPSS 23.0 software (IBM Corp., Armonk, NY, USA) was used for statistical analysis. Quantitative data are expressed as mean ± SD, single-factor analysis of variance was used for comparison between groups, and the Student–Newman–Keuls (SNK) method was used for pairwise comparison within groups. Qualitative data are expressed as percentages, and comparisons between groups were performed by χ2 test. A P value <0.05 was used to indicate statistically significant differences.
Results
Comparison of Kcr and H4ac between PCa tissues and adjacent tissues
As shown in Figure 1, the expression level of Kcr in PCa tissue was relatively weak, and it decreased to varying degrees in the nucleus and cytoplasm. The scores of the two modifications in PCa and adjacent tissues were also compared. The results showed that Kcr was significantly lower in PCa tissues than in adjacent tissues (P<0.05), whereas there was no statistical difference in the staining intensity and score of H4ac in cancerous versus adjacent tissues.

Correlation between Kcr and H4ac levels and PCa stages and grades
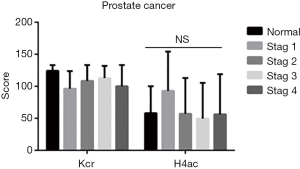
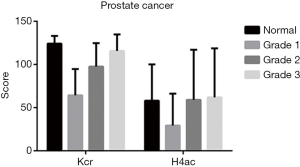
In PCa tissues of different clinical stages (stages 1–4), there was no difference between Kcr and H4ac (Figure 2). In terms of different grades (grades 1–3), the difference in H4ac levels was also not statistically significant. However, Kcr levels were found to increase with the increasing malignancy of PCa (Figure 3).
Evaluation of the effectiveness of BRD4 inhibitors in PCa cell lines
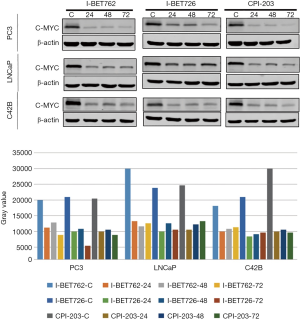
When the PC-3, LNCaP, and C42B cell lines were treated with I-BET762, I-BET726, and CPI-203, the expression level of C-MYC protein was measured, with the results showing that the expression level of C-MYC protein was significantly inhibited (Figure 4).
The effects of BRD4 inhibitors and sodium crotonate on the proliferation of PCa cell lines
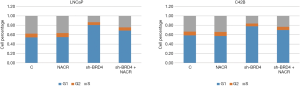
The results of flow cytometry measuring the cell cycles showed that after the addition of the three BRD4 inhibitors, the proportion of G1 phase cells of the PCa cell lines increased significantly, while the percentages of S phase cells decreased. However, when sodium crotonate was added, the cell cycle did not change significantly (Figures 5,6). CCK-8 testing results showed that the three BRD4 inhibitors had a significant inhibitory effect on the proliferation of PCa cell lines, whereas, after the addition of sodium crotonate, the cell proliferation function was significantly improved (Figures 7,8). Therefore, it seems that the BRD4 inhibitors could inhibit the proliferation of PCa cells, whereas histone crotonylation could promote the proliferation of PCa cell lines.

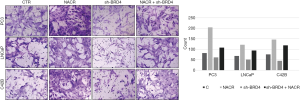
Effects of BRD4 inhibitors and sodium crotonate on the migration and invasion of PCa cell lines
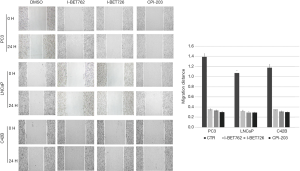
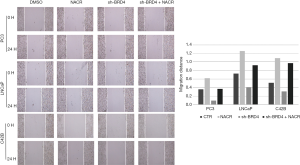
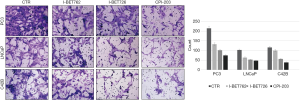
In the scratch test, the migration distance of PCa cell lines with the addition of BRD4 inhibitors was significantly shortened compared with that of the control group, whereas, when sodium crotonate was added, the migration distance of the PCa cell lines increased significantly compared to that of the control group (Figures 9,10). In the Transwell experiment, the number of PCa cells passing through the matrix membrane in the BRD4 inhibitor treatment groups was significantly reduced compared with that of the control group (Figures 11,12), while the addition of sodium crotonate significantly increased the number of cells passing through the matrix membrane. Therefore, BRD4 inhibitors could inhibit the migration and invasion of PCa cells, while histone crotonylation could promote the migration and invasion of PCa cells.
Effects of BRD4 inhibitors on histone modification
After the PC-3, LNCaP, and C42B cell lines were treated with I-BET762, I-BET726, and CPI-203 for 24, 48, and 72 h, the levels of histone modification of Kac, Kcr, and H3K18cr decreased significantly, and with the longer duration of treatment, both modifications showed a downward trend (Figure 13). To confirm that the decline of the two modifications was indeed caused by the inhibition of BRD4 function, we transiently knocked down the expression of BRD4 in three PCa cell lines and indeed found that the levels of histone modifications including those of Kac, Kcr, and H3K18cr were significantly downregulated (Figure 13) confirming that BRD4 could regulate histone modifications.
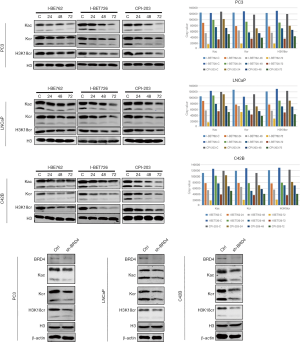
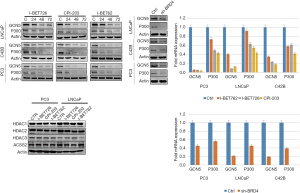
The effects of BRD4 inhibitors on histone modification-related enzymes
When PC-3, LNCaP, and C42B cell lines were treated with I-BET762, I-BET726, and CPI-203, the protein expression level of histone acetyltransferase GCN5 and P300 was downregulated; however, the protein expression level of histone deacetylase (HDAC) enzymes did not change (Figure 14). Furthermore, after BRD4 was transiently knocked down, the expression levels of those enzymes showed a similar pattern as that of the treatment groups (see Figure 14).
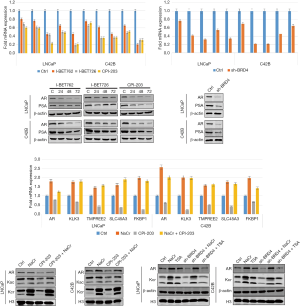
Effects of BRD4 inhibitors on androgen receptor signaling pathway
Since the androgen receptor signaling pathway plays an important role in the occurrence and development of PCa, we measured the expression level of marker proteins in this pathway. Since PC-3 cells are non–androgen-dependent cells, this experiment chose androgen-dependent PCa LNCaP cells and castration-resistant PCa C42B cells. After treating the two types of cells with I-BET762, I-BET726, and CPI-203, we found that the expression levels of androgen receptor and prostate-specific antigen decreased in the (Figure 15). Furthermore, we measured the expression levels of downstream target genes including AR, KLK3, TMPREE2, SLC45A3, and FKBP1 in the androgen receptor signaling pathway, and found that mRNA expression level was also significantly inhibited. Similar results were obtained when the BRD4 was knocked down transiently.
Discussion
The occurrence and development of PCa are the results of the malignant proliferation, migration, and invasion of tumor cells caused by a series of genetic and environmental factors; these factors constitute a complicated pathogenesis which eludes a comprehensive understanding (12). At present, the main treatment for PCa is surgery, radiotherapy, and chemotherapy. However, in the face of the diversity of PCa types, traditional treatment methods have encountered many insurmountable difficulties (13,14). In recent years, the studies on histone modification have progressed rapidly, and histone lysine residue can now be both acetylated and crotonylated. The structure of histone crotonylation modification is like that of acetylation modification, and it has a significant impact on the overall transcription functions (15,16). At present, relatively few studies have focused on the biological functions of histone crotonylation, and even fewer studies have examined its role in tumorigenesis and development. Since abnormalities of histone modification are common in tumors, we observed a high level of histone crotonylation in PCa tissues; furthermore, we found this level to be closely related to the pathologic grades of tumors. Therefore, we speculated that histone crotonylation may be a potential therapeutic target for PCa.
Hundreds of inhibitors for histone modification–related enzymes have been reported. These drugs can inhibit tumor proliferation, migration, invasion, differentiation, and epithelial–mesenchymal transition in different tumors (17). Moreover, many new high-efficiency small molecule inhibitors targeting effector proteins of histone acetylation have been developed, such as the BRD4 inhibitor, I-BET762 (3). BRD4 exists in multiple locations in the cell; it can undergo plasma–nuclear shuttling and affect the expression of cyclin E/D1, promoting cell proliferation and metastasis (18). Our study showed that BRD4 inhibitors have a significant inhibitory effect on the proliferation, migration, and invasion of PCa cells. Given its effectiveness on both non–androgen-dependent PC-3 and androgen castration-resistant C42B cells, this is of great significance to the treatments of these two currently difficult-to-treat PCa. Our results also showed that increasing histone crotonylation can promote the function of PCa cells, which is consistent with the observation that the increase of histone crotonylation in PCa correlated with the increasing malignancy.
Previous studies have shown that BRD4 is generally regarded as a recognition protein for histone acetylation modification and plays its biological function as an independent transcription factor (19). Other reports have also indicated that histones themselves can act as histone acetyltransferases to catalyze histone acetylation modification in vitro (6). Our results showed that BRD4 does not have the enzyme activity for histone modification; in other words, BRD4 cannot catalyze histone modification in vivo. However, it can regulate the level of histone modification by regulating the expression level of the histone acetylase, GCN5 and P300.
BRD4 can inhibit the activation of the androgen receptor signaling pathway, whereas histone crotonylation can activate the androgen receptor signaling pathway. By using BRD4 inhibitors and short hairpin RNA (sh-RNA) to inhibit and transiently knockdown BRD4 expression, we found that the expression level of androgen receptor and the activities of the androgen signaling pathway were inhibited. Furthermore, we also found that sodium crotonate could reactivate the androgen receptor signaling pathway by specifically improving the level of histone crotonylation. Importantly, this phenomenon was also observed in the androgen-resistant PCa cell line, C42B. It has been reported that mutations and re-splicing of the androgen receptor occur in castration-resistant PCa. The activation of abnormal androgen receptor signaling pathways is closely related to the progression, metastasis, invasion, and recurrence of PCa (20). Based on our results, we believe that BRD4 inhibitors can regulate histone crotonylation by regulating the expression of GCN5 and P300, thereby inhibiting the activation of the androgen receptor signaling pathway, which can potentially be used effectively in the treatment of castration-resistant PCa. Given the close relationship between histone crotonylation and PCa, we believe that the degree of malignancy of PCa is related to the excessive activation of the androgen receptor signaling pathway, although this needs to be further verified.
This study also has certain limitations. First, sodium crotonate can increase the level of the overall histone crotonylation in the cell. It is not clear which sites of the crotonylation cause the change of the biological function. This may require the use of antibodies targeting multiple sites of histone crotonylation coupled with chromatin immunoprecipitation (CHIP-Seq) to determine the corresponding biological functions for various histone site modifications. Secondly, recent studies have found that histone acetylases, GCN5 and P300, also have histone propionylase activity. The possibility that these enzymes have other functions cannot be ruled out, and thus the effects of other modifications are still not clear. In this study, we mainly focused on the expression of the androgen receptor signaling pathway, but whether or not histone crotonylation affects the mutation and splicing of the androgen receptor have not been studied. These all need to be further explored in subsequent research.
Conclusions
In PCa cell lines, histone acetylation effector protein BRD4 can regulate histone crotonylation by modulating the expression levels of histone acetyltransferases, GCN5 and P300. In androgen-dependent PCa cell line, LNCaP, and castration-resistant PCa cell line, C42B, histone crotonylation can specifically activate the androgen receptor signaling pathway. Histone crotonylation can promote cell proliferation, migration, and invasion of PCa cell lines. Therefore, histone crotonylation can be used as a potential therapeutic target for the treatment of PCa.
Acknowledgments
Funding: This work was supported by Scientific Research Project of Shanghai Municipal Health and Family Planning Commission (no. 201840245).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tau-21-53
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-21-53
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-21-53). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Greiman AK, Rosoff JS, Prasad SM. Association of Human Development Index with global bladder, kidney, prostate and testis cancer incidence and mortality. BJU Int 2017;120:799-807. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Asangani IA, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 2014;510:278-82. [Crossref] [PubMed]
- Lo EM, Balasubramanian A, Pastuszak AW, et al. Bipolar Androgen Therapy in Prostate Cancer (Update). J Sex Med 2020;17:831-4. [Crossref] [PubMed]
- Tan M, Luo H, Lee S, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011;146:1016-28. [Crossref] [PubMed]
- Zhao S, Zhang X, Li H. Beyond histone acetylation-writing and erasing histone acylations. Curr Opin Struct Biol 2018;53:169-77. [Crossref] [PubMed]
- Wei W, Liu X, Chen J, et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res 2017;27:898-915. [Crossref] [PubMed]
- Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer 2018;17:164. [Crossref] [PubMed]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014;54:728-36. [Crossref] [PubMed]
- Kanno T, Kanno Y, LeRoy G, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 2014;21:1047-57. [Crossref] [PubMed]
- Healy JR, Hart LS, Shazad AL, et al. Limited antitumor activity of combined BET and MEK inhibition in neuroblastoma. Pediatr Blood Cancer. 2020;67:e28267. [Crossref] [PubMed]
- Oudin MJ, Weaver VM. Physical and Chemical Gradients in the Tumor Microenvironment Regulate Tumor Cell Invasion, Migration, and Metastasis. Cold Spring Harb Symp Quant Biol 2016;81:189-205. [Crossref] [PubMed]
- Rim SH, Hall IJ, Massetti GM, et al. Primary Care Providers' Intended Use of Decision Aids for Prostate-Specific Antigen Testing for Prostate Cancer Screening. J Cancer Educ 2019;34:666-70. [Crossref] [PubMed]
- Mikuz G. Histologic classification of prostate cancer. Anal Quant Cytopathol Histpathol 2015;37:39-47. [PubMed]
- Wei W, Mao A, Tang B, et al. Large-Scale Identification of Protein Crotonylation Reveals Its Role in Multiple Cellular Functions. J Proteome Res 2017;16:1743-52. [Crossref] [PubMed]
- Sabari BR, Tang Z, Huang H, et al. Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation. Mol Cell 2018;69:533. [Crossref] [PubMed]
- Shah RR. Safety and Tolerability of Histone Deacetylase (HDAC) Inhibitors in Oncology. Drug Saf 2019;42:235-45. [Crossref] [PubMed]
- Liu Z, Tian B, Chen H, et al. Discovery of potent and selective BRD4 inhibitors capable of blocking TLR3-induced acute airway inflammation. Eur J Med Chem 2018;151:450-61. [Crossref] [PubMed]
- Chiang CM. Phospho-BRD4: transcription plasticity and drug targeting. Drug Discov Today Technol 2016;19:17-22. [Crossref] [PubMed]
- Loriot Y, Zoubeidi A, Gleave ME. Targeted therapies in metastatic castration-resistant prostate cancer: beyond the androgen receptor. Urol Clin North Am 2012;39:517-31. [Crossref] [PubMed]
(English Language Editor: J. Gray)




