Toward urinary cell-free DNA-based treatment of urothelial carcinoma: a narrative review
IntroductionOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Approximately 95% of urothelial carcinoma (UC) occurs in the lower urinary tract [urothelial bladder carcinoma (UBC)], whereas those in the upper urinary tract [upper tract urothelial carcinoma (UTUC)] are relatively rare (1).
UBC can be classified into two major groups: non-muscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC). Because NMIBC is associated with fewer deaths and a high risk of intravesical recurrence even after transurethral surgery, ranging from approximately 50% to 80% (2,3), patients with NMIBC require continuous follow-up with cystoscopy and urine cytology, making NMIBC one of the most prevalent and the most expensive cancer all over the world (4). For patients with localized MIBC, radical cystectomy, lymph node dissection, and cisplatin-based neoadjuvant chemotherapy (NAC) are the gold standard management proposed by current clinical guidelines (5-11). On the contrary, radical nephroureterectomy (RNU) with an ipsilateral bladder cuff and retroperitoneal lymph node dissection is one of the standard therapies for patients with muscle invasive, high-grade, or bulky UTUC, whereas kidney-sparing surgery (KSS) plays an increasingly important role in the management of low-risk UTUC, mainly due to improvements in endoscopic technologies (12). Even though the stage and grade of UTUC are decided by radiographical imaging or pathological tests obtained by endoscopic biopsy specimens, it is often difficult to determine accurately due to the thin wall of the upper urinary tract and the low volume of biopsy specimens (13). Because of the risk of recurrence during the follow-up period after RNU, surveillance based on cystoscopy, radiographical tests, and urinary cytology is mandatory. When KSS is performed, careful surveillance of the ipsilateral upper urinary tract is also necessary due to the high risk of local disease recurrence (12).
For these reasons, patients with UC, including UBC and UTUC, need to undergo several procedures at the time of initial diagnosis and continuous disease surveillance followed by initial therapy. Despite several commercially available assays for the detection of UC, such as UroVysion (14-20), urine cytology remains the most accessible and widely used noninvasive test worldwide. For the detection of UC, urine cytology established by The Paris System has high sensitivity for high-grade tumors (84%), but low sensitivity for low-grade tumors (16%) (21,22). Since current procedures for diagnosis or surveillance, such as cystoscopy or ureteroscopy (for patients after KSS) are invasive and costly, less invasive, and reliable follow-up methods are necessary for patients with (especially for low-grade) UC. Messer et al. reported that urine cytology is less sensitive in detecting UTUC than UBC and should be performed in selected cases of UTUC (23). In addition, there are no reliable prognostic biomarkers that reflect tumor grade or stage for patients who might need neoadjuvant or adjuvant chemotherapy. For these reasons, there is an urgent need to develop a noninvasive biomarker that could accurately diagnose, predict prognosis and response to drugs, and reveal the characteristics of the various stages of UC.
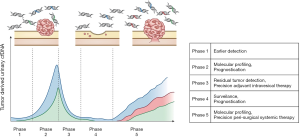
Liquid biopsy is a novel technology that detects and analyzes small amounts of tumor-derived cell-free DNA (cfDNA), extracellular vesicles (EVs), and circulating tumor cells (CTCs) in body fluids such as blood and urine (24). Recent technological innovations, such as the development of next-generation sequencing (NGS) or droplet digital PCR (ddPCR), have made it possible to perform molecular analysis of trace amounts of cell-free DNA in body fluids with high sensitivity. Liquid biopsy is expected to be indispensable for the realization of precision medicine (25,26). Owing to the fact that UC, unlike other carcinomas, is constantly in contact with urine, urinary cfDNA is an important source of genomic analysis of UC (27) (Figure 1). In this review, we summarize the current status, potential clinical applications, and limitations of cfDNA in urine as biomarkers in patients with UC. We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1259).
Literature search methodologyOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
We conducted a literature review of cfDNA about urothelial carcinoma. The literature was searched for publications up to May, 2020 for all English-language journals relating to cfDNA, using the PubMed database. The specific terms used were “bladder cancer” OR “bladder carcinoma” OR “urothelial cancer” OR “urothelial carcinoma” OR “upper tract” combined with “free DNA” OR “circulating DNA” OR “plasma DNA” OR “serum DNA” OR “urine DNA” OR “urinary DNA” OR “liquid biopsy” OR “liquid biopsies”.
Molecular biology of urothelial carcinoma tissueOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Due to technological advancements, NGS has made it possible to offer a comprehensive molecular characterization of various tumors, including UC. Over the last decade, molecular analysis of tumor specimens of UC has revealed details of genetic and epigenetic profiles and the association between molecular profiles and environmental factors. The classification of UC into molecular-based subtypes has facilitated more precisely decided treatment for patients with UC in addition to the prediction of prognosis, therapy resistance, and tumor progression (28-35).
Genetic analysisOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Using genome and transcriptome profiling, Robertson et al. demonstrated that the overall mutational burden in bladder cancer is associated with APOBEC-signature mutagenesis, and certain mutation signatures with a high-mutation subset are associated with better survival outcome in patients with MIBC (30). Hedegaard et al. also reported that the identification of molecular subtypes could offer better treatment strategies and better prognostication for patients with NMIBC (33). Hassler et al. compared the molecular characteristics of UBC and UTUC. Although UTUC and UBC share several similar genomic alterations, some molecular profiles are unique to UTUC-associated syndromes (34). Lynch syndrome-associated UTUC has germline mutations in mismatch repair genes or microsatellite instability. Aristolochic acid-induced UTUC has frequent A>T conversion or rare FGFR3 mutations. NGS analysis revealed several representatives of somatic mutations in UC. Mutations in the TERT promoter region are one of the most frequently occurring mutations in various neoplasms, including UC (36). Hotspot mutations in the TERT promoter mainly affect two regions, chromosome 5 of g.1295228 C>T and g.1295250 C>T, 124 and 146 upstream of bases of transcription start site (36,37). The mutant TERT promoter allele alters the binding capacity of transcription factors and engages in long-range chromatin interactions, subsequently stimulating TERT promoter activity and enabling tumors to overcome the end-replication problem and avoid senescence (38-40). TERT promoter mutations are widely identified in low-grade tumors, high-grade tumors, or rare histological variants (41-46). For these reasons, TERT promoter mutation is thought to be an early event in UC carcinogenesis (47).
PIK3CA is activated by point mutations at higher frequencies in NMIBC than in MIBC. PIK3CA mutations are most commonly associated with FGFR3 or RAS mutations. PIK3CA has domains for binding to RAS. There may be cooperation between PIK3CA mutant proteins and other events that are known to activate RAS (48).
RAS gene mutations (KRAS or HRAS mutations) are also frequently found in UBC. The Ras superfamily of monomeric G proteins has been reported to contribute to UBC progression with epidermal growth factor receptor, p53, and PTEN (49). TP53 mutations are rarely detected in low-grade UC, but are frequently detected in high-grade UC, including muscle invasive disease (30,33). TP53 mutations are also detected in carcinoma in situ (CIS) and are considered to be early events of CIS tumorigenesis. The primary function of TP53 genes is to control cell cycle progression, senescence, and cell death, leading to inhibition of tumorigenesis (50). These genes are activated by various cellular stresses, including DNA damage, hypoxia, replicative stress, and oncogene expression. A common cause of TP53 loss-of-function is an inactivating missense mutation. Fibroblast growth factor receptor 3 (FGFR3) is a glycoprotein composed of three extracellular immunoglobulin-like domains and a split tyrosine-kinase domain. An activating hotspot mutation in the FGFR3 gene or chromosomal translocation with a breakpoint near FGFR3 is frequently identified in UC (30,51). FGFR3 is characterized by a high prevalence of low stage tumors; these mutations are more frequently identified in pTa (80%) than in MIBC (10–20%). Given these facts, urinary cfDNA would help to differentiate low-grade tumors from high-grade tumors by analyzing TP53 and FGFR3 mutations.
Epigenetic analysisOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
In a previous study, Wolff et al. observed that 12% of all CpG loci were hypermethylated in bladders with cancer (52). They also reported that carcinogen exposure rather than a clonal expansion seems to result in these epigenetic alterations. DNA methylation is the most studied epigenetic modification in cancer and is thought to be a prevalent and early event in tumorigenesis (53). Hypermethylation of CpG islands represses tumor suppressor gene transcription either directly by inhibiting transcription factor binding or indirectly through the recruitment of methyl-CpG-binding proteins. Hypomethylated CpG regions might activate oncogenes, resulting in chromosome instability (54,55). Although methylation-specific PCR (MSP) is a sensitive method for detecting a methylated region of interest using specific primers on bisulfite-converted DNA, recent technological advancements in bisulfite sequencing have made it possible to analyze methylation of DNA on a genome-wide scale (56). These genome-wide methylation analyses will lead to a deeper understanding of how the genetic function is implemented and regulated. Epigenetic alterations in the promoter regions of susceptible genes can be detected in both blood and urinary cfDNA, including key cellular pathways such as cell cycle control, apoptosis, cell differentiation, DNA repair, cellular adhesion, and migration.
Overview of the cell-free DNAOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
The cfDNA released into body fluids by apoptosis, necrosis, and secretion of both cancerous and noncancerous tissues is free from any type of cells as fragmented nucleic acids (24). The presence of tumor-derived cfDNA in serum was first reported by Leon et al. in 1977 (57). Due to the minimal amount of cfDNA in body fluids and the requirement of a sensitive assay for analysis, the research in this field was limited for a while. In 1997, Lo et al. reported for the first time that fetal cfDNA circulates in maternal plasma (58), which led to the development of noninvasive prenatal genetic testing. Advances in obstetrics and genetic analysis techniques have led to rapid progress in research on tumor-derived cfDNA.
The analysis of cfDNA has several advantages over conventional tissue biopsy. It allows us to analyze in a less invasive, rapid, repeated, sequential manner, and also to overcome the heterogeneity of tumors (59). Thus, cfDNA can provide useful genetic information and offer several advantages not only as a noninvasive diagnostic tool, but also as an early indicator of minimal residual disease, recurrence, drug resistance, or metastasis, and believed to replace tissue biopsy in the near future.
Although cfDNA can be detected in both plasma and serum, plasma samples are preferred over serum for cfDNA analysis because blood cell lysis during the preparation of serum samples could release DNA from noncancerous cells, which might lower the sensitivity of cfDNA detection derived from tumors (60).
However, limited DNA quantity in plasma makes it difficult to detect mutations with very low variant allele frequency or those of localized tumors because the amount of cfDNA in plasma is mainly correlated with tumor burden in various types of malignancies. For these reasons, the majority of previous studies focused on advanced tumor stages, and the clinical utility of cfDNA mutational analysis in blood for the detection of early-stage tumors is less well documented (61). Still, owing to the fact that UC, unlike other carcinomas, is in constant contact with urine, urinary cfDNA is an important source of genomic analysis of UC (27).
DNA sources in urine consist of both urinary sedimented cells (pellet DNA) and urinary supernatant (cfDNA) separated by centrifugation, and the utility of pellet DNA analysis has long been established (62,63). Importantly, urinary cfDNA in the supernatant, discovered later than the pellet, has been reported to exhibit a higher sensitivity than the pellet DNA in several papers (64-66). Urinary cfDNA can be classified into two groups according to their fragment size: high-molecular-weight (1 kbp or longer) and low-molecular-weight (67,68). High-molecular-weight urinary cfDNA is derived from necrotic cells or lymphocytes in the urinary tract (67,69). However, low-molecular-weight urinary cfDNA can originate either from apoptotic cells or systemic circulation (67,70). Bryzgunova et al. estimated that more than 3×106 epithelial cells could be excreted into the urine in one day, and some of these epithelial cells undergo apoptosis to release cfDNA into the urine (71). Furthermore, urinary cfDNA carries genetic information of circulating cfDNA that passes through the glomerular barrier, known as trans-renal DNA (69). For example, EGFR mutations have been detected in the urinary cfDNA of patients with non-small-cell lung cancer (71), and KRAS mutations have been detected in patients with colorectal cancer (66,72) and pancreatic cancer (73). More importantly, urine has the advantage of being a noninvasive sample source over tissue and blood. Urine can be collected in cups without any specialized skills or equipment.
Techniques for efficient detection of urinary cfDNAOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
In order to detect urinary cfDNA efficiently, some techniques are essential for urine storage, isolation of urinary cfDNA, and detection method. Since voided urine contains a lot of contaminants like bacteria, blood cells, or crystals, it is necessary to exclude artifacts derived from these contaminants as much as possible. Because voided urine samples from patients are susceptible to crystal precipitation even after refrigeration at 2 to 8 °C for several hours, urine sample processing, including centrifugation and removal of crystals and cellular components followed by storage at −80 °C, should be performed promptly after urine collection (74,75). Although cfDNA can be isolated by classical laboratory techniques, many types of commercial kits are available for the isolation of cfDNA. For the detection of genomic alterations efficiently, NGS and ddPCR technology are widely used. Each of these methods has its own advantages and features that need to be well understood. NGS is a high-powered analysis method capable of running millions of sequences simultaneously, enabling comprehensive detection of gene mutations. Furthermore, ddPCR allows for highly accurate measurements of DNA content by dividing a nucleic acid sample into thousands of parallel PCR reactions, each of which is read separately and analyzed statistically. Since the ddPCR technique is based on a given specific primer, it is not suitable for extensive exploratory measurements; however, it offers higher accuracy and sensitivity than other methods.
Association of genomic alteration profiles between tumor tissue and cfDNAOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Several researchers have investigated the concordance of genomic alteration profiles between tumor tissue and cfDNA. A previous study by Agarwal et al. analyzed the comprehensive genomic profile of plasma cfDNA in metastatic UC, including UTUC, and compared the frequency of genomic mutations between tumor tissue and cfDNA (76). They observed a similar frequency of recurrent mutations (TP53, ARID1A, PIK3CA, ERBB2, and FGFR3) between tumor tissues of patients with MIBC and cfDNA of patients with metastatic UBC. Dudley et al. observed a certain concordance of mutations in tumor tissue and urinary cfDNA obtained from paired urine and tissue samples of 18 patients with UBC. They also reported that tumor mutations also identified in urine had a higher median allele fraction (27% vs. 9%, P<0.0001) than those not identified in urine, indicating that genomic concordance between mutations found in tumor tissue and urinary cfDNA is higher for truncal mutations (77). Cheng et al. investigated the genome-wide analysis of methylation status of urinary cfDNA in patients with UBC. Shallow-depth paired-end genome-wide bisulfite sequencing revealed that the tumor tissue and urinary cfDNA have a significant positive correlation in methylomes (78).
In short, there seems to be an overall concordance between tumor tissue and cfDNA in terms of genomic alterations, including genomic mutations and methylation alterations.
Clinical application of cfDNA analysis for patients with UBCOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Several methods for cfDNA analysis have been reported. In the pre-NGS era, when devices such as NGS or ddPCR, which can detect a small number of mutated alleles, were not prevalent, microsatellite, methylation, or quantification of blood cfDNA were mainly investigated for clinical use.
Quantification of cfDNAOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Several studies have demonstrated the quantification of cfDNA. Brisuda et al. reported that urinary cfDNA levels in patients with UBC were significantly higher than those in the control group, with a sensitivity of 42.4% and a specificity of 91.2%. However, the levels of urinary cfDNA are affected by hematuria or pyuria, and attention is necessary for clinical use (79).
Microsatellite analysisOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Microsatellite analysis was performed in the early era of cfDNA investigation. Microsatellites are highly polymorphic DNA-repeat regions, and alterations of microsatellite DNA are thought to be valuable markers for detecting UC (80,81). Utting et al. reported that multiple simultaneous investigations of microsatellite markers of urinary and plasma cfDNA have clinical potential for noninvasive diagnosis of UBC (82). Szarvas et al. reported that microsatellite analysis of urinary cfDNA is more sensitive for the detection of UBC than that of urine pellet DNA (66).
Integrity of cfDNAOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
It has been reported that cfDNA from apoptotic cells is highly fragmented, whereas cfDNA from necrotic cells results in longer DNA fragments. Casadio et al. reported that the DNA integrity of urinary cfDNA showed a sensitivity of 73% and a specificity of 83% in symptomatic patients. In their report, urinary cfDNA integrity was defined as the quantity of c-Myc, BCAS1, and HER2 with sequences longer than 250 bp by real time PCR (83).
Epigenetic alterations in cfDNAOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
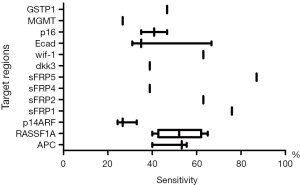
To date, there have been few reports on the epigenetic analysis of urinary “cfDNA” (Table 1) (78,84,85), but many reports on urinary “pellet” DNA (86-101) using various ranges of methylation biomarkers. The combination of multiple methylation levels showed a high sensitivity for UBC diagnosis. However, the sensitivity and specificity of each single methylation marker differ across several studies (Figure 2). Commercially available AssureMDx (MDx Health, USA) evaluated the DNA methylation of OTX1, ONECUT2, and TWIST and mutation analysis of FGFR3, TERT promoter, and HRAS in urinary pellet DNA for the detection of UC (101). The sensitivity and specificity of AssureMDx were 97% and 83% in 74 patients with UBC and 80 patients with gross hematuria, respectively. Because hematuria is an important symptom of UC, this commercial assay has great potential for clinical use, reducing unnecessary cystoscopies in patients with hematuria. In comparison with studies on epigenomics in urinary pellet DNA, few reports have been made about urinary cfDNA. Cheng et al. reported high sensitivity and specificity of shallow-depth genome-wide bisulfite sequencing of urinary cfDNA (78). In serum cfDNA, hypermethylation of 3 genes (APC, GSTP1, and TIG1) provided not only diagnostic but also valuable prognostic information for patients with UBC (102). Others reported that the methylation levels of p16 and DAPK can be used to differentiate NMIBC from healthy controls with 76.2% sensitivity and 100% specificity (103). Furthermore, the point that DAPK promoter methylation was detected in the serum of 71.4% of patients with low-grade NMIBC is worth noting. Thus, a unique feature of the methylation assay is that it can be performed on blood samples, even in patients with early stages of UC. However, there are certain issues with methylation analysis that need to be solved. First, there is great variability in the positive rate in each study. Second, unlike genetic mutations, the changes are not concentrated in a specific locus, and last, considerable DNA losses occur under bisulfite treatment.

Full table
Genetic alterations in the cfDNAOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
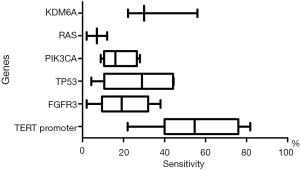
In order to detect low volumes of mutant DNA in urine, targeted panel methods are often used to ensure the accuracy and efficiency of genetic analysis of DNA in urine (77,104). There have been several reports about the clinical utility of urinary cfDNA in patients with UC (64,65,77,105-116) (Table 2). Because genetic alterations are highly specific to cancer, it is possible to increase the sensitivity of UBC detection by examining many mutations while maintaining specificity (77,105). For this reason, it is necessary to balance the cost by determining the number of genetic mutations to be analyzed. Due to the limited number of frequent genetic alterations in UBC, more genetic alteration data on urinary cfDNA have been accumulated from various institutions at various stages of cancer than that of epigenome analysis (Figure 3).
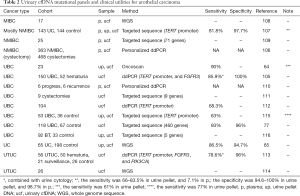
Full table
Dudley et al. used high-throughput targeted sequence CAPP-Seq assay to detect frequent mutations in urinary cfDNA for diagnosis and surveillance of UBC (77). The sensitivity of CAPP-Seq was 84–93%, and the specificity was 96–100%. They also reported that this targeted sequence could detect 91% of UBC recurrence in the surveillance setting, and significantly outperformed current clinical tests such as UroVysion or urine cytology. Christensen et al. reported the clinical utility of personalized cfDNA analysis for disease surveillance of patients with UBC (106). They developed urine and plasma cfDNA hotspot mutational assays by ddPCR for patients with FGFR3 and PIK3CA tumor mutations, and reported that these mutations in urinary cfDNA were significantly associated with disease progression from NMIBC. Furthermore, a positive correlation of tumor-derived DNA between urine and plasma was observed. We have reported the utility of ddPCR-based simple urinary cfDNA assay targeting 3 mutation sites (2 promoter mutations of TERT and 1 FGFR3 mutation) (105). In this report, the sensitivity was 77.5% to 85.9% in conjunction with urine cytology, and the specificity was 100%. Furthermore, in disease surveillance after transurethral resection of bladder tumor, patients with mutant TERT C228T positive urinary cfDNA had a significantly worse prognosis for bladder tumor recurrence than patients with negative mutations. Since TERT promoter mutations are frequently detected in UC, TERT promoter mutation is thought to be one of the strong candidates for noninvasive biomarkers in urinary cfDNA in many reports (105,107,112,115).
Clinical application of cfDNA analysis in UTUCOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Due to the small number of patients with UTUC, there have been few reports on urinary cfDNA analysis in UTUC. We investigated the diagnostic and prognostic potential of urinary cfDNA in patients with localized UTUC (113). In our report, the sensitivity of 3 hotspot gene mutations in urinary cfDNA was only 55.4%, but the sensitivity increased to 78.6% when combined with urine cytology. Interestingly, urinary cfDNA collected after radical nephroureterectomy was a predictor of bladder tumor recurrence after radical surgery. Lu et al. investigated the whole genome sequence of urinary cfDNA in Chinese patients with UTUC (114). They concluded that a mutational signature in urinary cfDNA induced by aristolochic acid might serve as a screening tool to define low-risk UTUC with therapeutic relevance. Furthermore, this mutational signature in urinary cfDNA is useful for diagnostic uncertainty when kidney-sparing treatment and/or systemic therapy are considered.
Challenges and future directions for urinary cfDNA analysisOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Current diagnostic procedures have some issues to be solved. First, urine cytology has low sensitivity for low-grade tumor; second, urinary tract endoscopy is invasive; third, BCG-induced inflammatory changes in the urothelium are often mistaken for CIS or other malignant manifestations on cystoscopy. Urinary cfDNA analysis can compensate for the weaknesses of current procedures and strengthen diagnostic capabilities, as proposed by many guidelines. It is necessary to conduct prospective multi-institutional clinical trials that address specific clinical questions in an accurate and precise manner because current reports about urinary cfDNA are mainly based on a small number of patients in a single institute. Urine cytology has a high specificity and a long history of clinical use. We believe that in the future, urinary liquid biopsy can help in improving diagnosis by combining morphological evaluation of urine cytology and molecular assessment of urinary cfDNA. TERT promoter mutations have been detected in the non-malignant urothelium of patients with UBC, and these mutations are significantly associated with bladder tumor recurrence (117). The fact that urinary cfDNA alterations are detected in patients without visible tumor indicates that these mutated cfDNAs originate from non-malignant urothelium, reflecting the whole bladder urothelium genomic status. Though this might reduce positive predictive value, patients with mutated cfDNA in urine without visible tumor have a high likelihood of developing tumors and should be followed carefully. Furthermore, it might be useful for patients with MIBC after bladder preservation therapy by urinary cfDNA analysis. Urinary cfDNA analysis may offer not only a novel initial workup for patients suspected of having UC, but also a novel follow-up strategy or novel adjuvant therapy for patients in the surveillance.
AcknowledgmentsOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Funding: None.
FootnoteOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Andrology and Urology for the series “Urinary Biomarkers of Urothelial Malignancies”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the NARRATIVE REVIEW reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1259
Peer Review File: Available at http://dx.doi.org/10.21037/tau-20-1259
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1259). The series “Urinary Biomarkers of Urothelial Malignancies” was commissioned by the editorial office without any funding or sponsorship. KF serves as an unpaid editorial board member of Translational Andrology and Urology from Oct 2019 to Sep 2021 and served as the unpaid Guest Editor of the series. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Introduction
- Literature search methodology
- Molecular biology of urothelial carcinoma tissue
- Genetic analysis
- Epigenetic analysis
- Overview of the cell-free DNA
- Techniques for efficient detection of urinary cfDNA
- Association of genomic alteration profiles between tumor tissue and cfDNA
- Clinical application of cfDNA analysis for patients with UBC
- Quantification of cfDNA
- Microsatellite analysis
- Integrity of cfDNA
- Epigenetic alterations in cfDNA
- Genetic alterations in the cfDNA
- Clinical application of cfDNA analysis in UTUC
- Challenges and future directions for urinary cfDNA analysis
- Acknowledgments
- Footnote
- References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Prout GR Jr, Barton BA, Griffin PP, et al. Treated history of noninvasive grade 1 transitional cell carcinoma. The National Bladder Cancer Group. J Urol 1992;148:1413-9. [Crossref] [PubMed]
- Sylvester RJ, van der Meijden AP, Oosterlinck W, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol 2006;49:466-5; discussion 475-7. [Crossref] [PubMed]
- Svatek RS, Hollenbeck BK, Holmang S, et al. The economics of bladder cancer: costs and considerations of caring for this disease. Eur Urol 2014;66:253-62. [Crossref] [PubMed]
- Stein JP, Lieskovsky G, Cote R, et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol 2001;19:666-75. [Crossref] [PubMed]
- Stein JP, Skinner DG. Radical cystectomy for invasive bladder cancer: long-term results of a standard procedure. World J Urol 2006;24:296-304. [Crossref] [PubMed]
- Dalbagni G, Genega E, Hashibe M, et al. Cystectomy for bladder cancer: a contemporary series. J Urol 2001;165:1111-6. [Crossref] [PubMed]
- Bassi P, Ferrante GD, Piazza N, et al. Prognostic factors of outcome after radical cystectomy for bladder cancer: a retrospective study of a homogeneous patient cohort. J Urol 1999;161:1494-7. [Crossref] [PubMed]
- Ghoneim MA, el-Mekresh MM, el-Baz MA, et al. Radical cystectomy for carcinoma of the bladder: critical evaluation of the results in 1,026 cases. J Urol 1997;158:393-9. [Crossref] [PubMed]
- David KA, Milowsky MI, Ritchey J, et al. Low incidence of perioperative chemotherapy for stage III bladder cancer 1998 to 2003: a report from the National Cancer Data Base. J Urol 2007;178:451-4. [Crossref] [PubMed]
- Porter MP, Kerrigan MC, Donato BM, et al. Patterns of use of systemic chemotherapy for Medicare beneficiaries with urothelial bladder cancer. Urol Oncol 2011;29:252-8. [Crossref] [PubMed]
- Seisen T, Peyronnet B, Dominguez-Escrig JL, et al. Oncologic Outcomes of Kidney-sparing Surgery Versus Radical Nephroureterectomy for Upper Tract Urothelial Carcinoma: A Systematic Review by the EAU Non-muscle Invasive Bladder Cancer Guidelines Panel. Eur Urol 2016;70:1052-68. [Crossref] [PubMed]
- Smith AK, Stephenson AJ, Lane BR, et al. Inadequacy of biopsy for diagnosis of upper tract urothelial carcinoma: implications for conservative management. Urology 2011;78:82-6. [Crossref] [PubMed]
- Beukers W, van der Keur KA, Kandimalla R, et al. FGFR3, TERT and OTX1 as a Urinary Biomarker Combination for Surveillance of Patients with Bladder Cancer in a Large Prospective Multicenter Study. J Urol 2017;197:1410-8. [Crossref] [PubMed]
- Critelli R, Fasanelli F, Oderda M, et al. Detection of multiple mutations in urinary exfoliated cells from male bladder cancer patients at diagnosis and during follow-up. Oncotarget 2016;7:67435-48. [Crossref] [PubMed]
- van der Aa MN, Zwarthoff EC, Steyerberg EW, et al. Microsatellite analysis of voided-urine samples for surveillance of low-grade non-muscle-invasive urothelial carcinoma: feasibility and clinical utility in a prospective multicenter study (Cost-Effectiveness of Follow-Up of Urinary Bladder Cancer trial [CEFUB]). Eur Urol 2009;55:659-67. [Crossref] [PubMed]
- Rouprêt M, Hupertan V, Yates DR, et al. A comparison of the performance of microsatellite and methylation urine analysis for predicting the recurrence of urothelial cell carcinoma, and definition of a set of markers by Bayesian network analysis. BJU Int 2008;101:1448-53. [Crossref] [PubMed]
- Todenhöfer T, Hennenlotter J, Guttenberg P, et al. Prognostic relevance of positive urine markers in patients with negative cystoscopy during surveillance of bladder cancer. BMC Cancer 2015;15:155. [Crossref] [PubMed]
- Grossman HB, Messing E, Soloway M, et al. Detection of bladder cancer using a point-of-care proteomic assay. JAMA 2005;293:810-6. [Crossref] [PubMed]
- Kim PH, Sukhu R, Cordon BH, et al. Reflex fluorescence in situ hybridization assay for suspicious urinary cytology in patients with bladder cancer with negative surveillance cystoscopy. BJU Int 2014;114:354-9. [PubMed]
- Yafi FA, Brimo F, Steinberg J, et al. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol Oncol 2015;33:66.e25 [Crossref] [PubMed]
- Allison DB, VandenBussche CJ. A Review of Urine Ancillary Tests in the Era of the Paris System. Acta Cytol 2020;64:182-92. [Crossref] [PubMed]
- Messer J, Shariat SF, Brien JC, et al. Urinary cytology has a poor performance for predicting invasive or high-grade upper-tract urothelial carcinoma. BJU Int 2011;108:701-5. [Crossref] [PubMed]
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Maia MC, Salgia M, Pal SK. Harnessing cell-free DNA: plasma circulating tumour DNA for liquid biopsy in genitourinary cancers. Nat Rev Urol 2020;17:271-91. [Crossref] [PubMed]
- Valenberg FJPV, Hiar AM, Wallace E, et al. Validation of an mRNA-based Urine Test for the Detection of Bladder Cancer in Patients with Haematuria. Eur Urol Oncol 2021;4:93-101. [Crossref] [PubMed]
- Hayashi Y, Fujita K. A new era in the detection of urothelial carcinoma by sequencing cell-free DNA. Transl Androl Urol 2019;8:S497-501. [Crossref] [PubMed]
- Dyrskjøt L, Thykjaer T, Kruhøfferr M, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet 2003;33:90-6. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171:540-56.e25. [Crossref] [PubMed]
- Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 2011;43:875-8. [Crossref] [PubMed]
- Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014;25:152-65. [Crossref] [PubMed]
- Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016;30:27-42. [Crossref] [PubMed]
- Hassler MR, Bray F, Catto JWF, et al. Molecular Characterization of Upper Tract Urothelial Carcinoma in the Era of Next-generation Sequencing: A Systematic Review of the Current Literature. Eur Urol 2020;78:209-220. [Crossref] [PubMed]
- Hayashi T, Fujita K, Hayashi Y, et al. Mutational Landscape and Environmental Effects in Bladder Cancer. Int J Mol Sci 2020;21:E6072 [Crossref] [PubMed]
- Vinagre J, Almeida A. Frequency of TERT promoter mutations in human cancers. Nat Commun 2013;4:2185. [Crossref] [PubMed]
- Kinde I, Munari E, Faraj SF, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res 2013;73:7162-7. [Crossref] [PubMed]
- Stern JL, Theodorescu D, Vogelstein B, et al. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev 2015;29:2219-24. [Crossref] [PubMed]
- Min J, Shay JW. TERT Promoter Mutations Enhance Telomerase Activation by Long-Range Chromatin Interactions. Cancer Discov 2016;6:1212-4. [Crossref] [PubMed]
- Borah S, Xi L, Zaug AJ, et al. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 2015;347:1006-10. [Crossref] [PubMed]
- Rodriguez Pena MDC, Tregnago AC, Eich ML, et al. Spectrum of genetic mutations in de novo PUNLMP of the urinary bladder. Virchows Arch 2017;471:761-7. [Crossref] [PubMed]
- Nguyen D, Taheri D, Springer S, et al. High prevalence of TERT promoter mutations in micropapillary urothelial carcinoma. Virchows Arch 2016;469:427-34. [Crossref] [PubMed]
- Cowan ML, Springer S, Nguyen D, et al. Detection of TERT promoter mutations in primary adenocarcinoma of the urinary bladder. Hum Pathol 2016;53:8-13. [Crossref] [PubMed]
- Cowan M, Springer S, Nguyen D, et al. High prevalence of TERT promoter mutations in primary squamous cell carcinoma of the urinary bladder. Mod Pathol 2016;29:511-5. [Crossref] [PubMed]
- Zheng X, Zhuge J, Bezerra SM, et al. High frequency of TERT promoter mutation in small cell carcinoma of bladder, but not in small cell carcinoma of other origins. J Hematol Oncol 2014;7:47. [Crossref] [PubMed]
- Palsgrove DN, Taheri D, Springer SU, et al. Targeted sequencing of plasmacytoid urothelial carcinoma reveals frequent TERT promoter mutations. Hum Pathol 2019;85:1-9. [Crossref] [PubMed]
- Chiba K, Lorbeer FK, Shain AH, et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science 2017;357:1416-20. [Crossref] [PubMed]
- Knowles MA, Platt FM, Ross RL, et al. Phosphatidylinositol 3-kinase (PI3K) pathway activation in bladder cancer. Cancer Metastasis Rev 2009;28:305-16. [Crossref] [PubMed]
- Oxford G, Theodorescu D. The role of Ras superfamily proteins in bladder cancer progression. J Urol 2003;170:1987-93. [Crossref] [PubMed]
- Bykov VJN, Eriksson SE, Bianchi J, et al. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer 2018;18:89-102. [Crossref] [PubMed]
- Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013;22:795-803. [Crossref] [PubMed]
- Wolff EM, Chihara Y, Pan F, et al. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res 2010;70:8169-78. [Crossref] [PubMed]
- Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev 1988;2:1136-43. [Crossref] [PubMed]
- Eden A, Gaudet F, Waghmare A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003;300:455. [Crossref] [PubMed]
- Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science 2003;300:489-92. [Crossref] [PubMed]
- Gupta R, Nagarajan A, Wajapeyee N. Advances in genome-wide DNA methylation analysis. Biotechniques 2010;49:iii-xi. [Crossref] [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed]
- Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997;350:485-7. [Crossref] [PubMed]
- Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed?. J Clin Oncol 2013;31:17-22. [Crossref] [PubMed]
- El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013;424:222-30. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Lu T, Li J. Clinical applications of urinary cell-free DNA in cancer: current insights and promising future. Am J Cancer Res 2017;7:2318-32. [PubMed]
- Zeng S, Ying Y, Xing N, et al. Noninvasive Detection of Urothelial Carcinoma by Cost-effective Low-coverage Whole-genome Sequencing from Urine-Exfoliated Cell DNA. Clin Cancer Res 2020;26:5646-54. [Crossref] [PubMed]
- Togneri FS, Ward DG, Foster JM, et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur J Hum Genet 2016;24:1167-74. [Crossref] [PubMed]
- Ge G, Peng D, Guan B, et al. Urothelial Carcinoma Detection Based on Copy Number Profiles of Urinary Cell-Free DNA by Shallow Whole-Genome Sequencing. Clin Chem 2020;66:188-98. [Crossref] [PubMed]
- Szarvas T, Kovalszky I, Bedi K, et al. Deletion analysis of tumor and urinary DNA to detect bladder cancer: urine supernatant versus urine sediment. Oncol Rep 2007;18:405-9. [Crossref] [PubMed]
- Su YH, Wang M, Brenner DE, et al. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J Mol Diagn 2004;6:101-7. [Crossref] [PubMed]
- Bryzgunova OE, Skvortsova TE, Kolesnikova EV, et al. Isolation and comparative study of cell-free nucleic acids from human urine. Ann N Y Acad Sci 2006;1075:334-40. [Crossref] [PubMed]
- Su YH, Wang M, Block TM, et al. Transrenal DNA as a diagnostic tool: important technical notes. Ann N Y Acad Sci 2004;1022:81-9. [Crossref] [PubMed]
- Salvi S, Martignano F, Molinari C, et al. The potential use of urine cell free DNA as a marker for cancer. Expert Rev Mol Diagn 2016;16:1283-90. [Crossref] [PubMed]
- Bryzgunova OE, Laktionov PP. Extracellular Nucleic Acids in Urine: Sources, Structure Diagnostic Potential. Acta Naturae 2015;7:48-54. [Crossref] [PubMed]
- Sands J, Li Q, Hornberger J. Urine circulating-tumor DNA (ctDNA) detection of acquired EGFR T790M mutation in non-small-cell lung cancer: An outcomes and total cost-of-care analysis. Lung Cancer 2017;110:19-25. [Crossref] [PubMed]
- Botezatu I, Serdyuk O, Potapova G, et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem 2000;46:1078-84. [Crossref] [PubMed]
- Xia Y, Huang CC, Dittmar R, et al. Copy number variations in urine cell free DNA as biomarkers in advanced prostate cancer. Oncotarget 2016;7:35818-31. [Crossref] [PubMed]
- Riberiro KCB, Serabion BRL, Nolasco EL, et al. Urine storage under refrigeration preserves the sample in chemical, cellularity and bacteriuria analysis of ACS. J Bras Patol Med Lab 2013;49:415-22. [Crossref]
- Agarwal N, Pal SK, Hahn AW, et al. Characterization of metastatic urothelial carcinoma via comprehensive genomic profiling of circulating tumor DNA. Cancer 2018;124:2115-24. [Crossref] [PubMed]
- Dudley JC, Schroers-Martin J, Lazzareschi DV, et al. Detection and Surveillance of Bladder Cancer Using Urine Tumor DNA. Cancer Discov 2019;9:500-9. [Crossref] [PubMed]
- Cheng THT, Jiang P, Teoh JYC, et al. Noninvasive Detection of Bladder Cancer by Shallow-Depth Genome-Wide Bisulfite Sequencing of Urinary Cell-Free DNA for Methylation and Copy Number Profiling. Clin Chem 2019;65:927-36. [Crossref] [PubMed]
- Brisuda A, Pazourkova E, Soukup V, et al. Urinary Cell-Free DNA Quantification as Non-Invasive Biomarker in Patients with Bladder Cancer. Urol Int 2016;96:25-31. [Crossref] [PubMed]
- Vieira ML, Santini L, Diniz AL, et al. Microsatellite markers: what they mean and why they are so useful. Genet Mol Biol 2016;39:312-28. [Crossref] [PubMed]
- Steiner G, Schoenberg MP, Linn JF, et al. Detection of bladder cancer recurrence by microsatellite analysis of urine. Nat Med 1997;3:621-4. [Crossref] [PubMed]
- Utting M, Werner W, Dahse R, et al. Microsatellite analysis of free tumor DNA in urine, serum, and plasma of patients: a minimally invasive method for the detection of bladder cancer. Clin Cancer Res 2002;8:35-40. [PubMed]
- Casadio V, Calistri D, Tebaldi M, et al. Urine cell-free DNA integrity as a marker for early bladder cancer diagnosis: preliminary data. Urol Oncol 2013;31:1744-50. [Crossref] [PubMed]
- Roperch JP, Hennion C. A novel ultra-sensitive method for the detection of FGFR3 mutations in urine of bladder cancer patients - Design of the Urodiag PCR kit for surveillance of patients with non-muscle-invasive bladder cancer (NMIBC). BMC Med Genet 2020;21:112. [Crossref] [PubMed]
- Hentschel AE, Nieuwenhuijzen JA, Bosschieter J, et al. Comparative Analysis of Urine Fractions for Optimal Bladder Cancer Detection Using DNA Methylation Markers. Cancers (Basel) 2020;12:859. [Crossref] [PubMed]
- Dulaimi E, Uzzo RG, Greenberg RE, et al. Detection of bladder cancer in urine by a tumor suppressor gene hypermethylation panel. Clin Cancer Res 2004;10:1887-93. [Crossref] [PubMed]
- Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst 2006;98:996-1004. [Crossref] [PubMed]
- Urakami S, Shiina H, Enokida H, et al. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res 2006;12:2109-16. [Crossref] [PubMed]
- Yates DR, Rehman I, Meuth M, et al. Methylational urinalysis: a prospective study of bladder cancer patients and age stratified benign controls. Oncogene 2006;25:1984-8. [Crossref] [PubMed]
- Lin HH, Ke HL, Huang SP, et al. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of E-cadherin, p16, p14, RASSF1A genes in urine. Urol Oncol 2010;28:597-602. [Crossref] [PubMed]
- Zhao Y, Guo S, Sun J, et al. Methylcap-seq reveals novel DNA methylation markers for the diagnosis and recurrence prediction of bladder cancer in a Chinese population. PLoS One 2012;7:e35175 [Crossref] [PubMed]
- Beukers W, Kandimalla R, van Houwelingen D, et al. The use of molecular analyses in voided urine for the assessment of patients with hematuria. PLoS One 2013;8:e77657 [Crossref] [PubMed]
- Zuiverloon TC, Beukers W, van der Keur KA, et al. Combinations of urinary biomarkers for surveillance of patients with incident nonmuscle invasive bladder cancer: the European FP7 UROMOL project. J Urol 2013;189:1945-51. [Crossref] [PubMed]
- Andersson E, Dahmcke CM, Steven K, et al. Filtration Device for On-Site Collection, Storage and Shipment of Cells from Urine and Its Application to DNA-Based Detection of Bladder Cancer. PLoS One 2015;10:e0131889 [Crossref] [PubMed]
- Roperch JP, Grandchamp B, Desgrandchamps F, et al. Promoter hypermethylation of HS3ST2, SEPTIN9 and SLIT2 combined with FGFR3 mutations as a sensitive/specific urinary assay for diagnosis and surveillance in patients with low or high-risk non-muscle-invasive bladder cancer. BMC Cancer 2016;16:704. [Crossref] [PubMed]
- Dahmcke CM, Steven KE, Larsen LK, et al. A Prospective Blinded Evaluation of Urine-DNA Testing for Detection of Urothelial Bladder Carcinoma in Patients with Gross Hematuria. Eur Urol 2016;70:916-9. [Crossref] [PubMed]
- Tan WS, Feber A, Dong L, et al. DETECT I & DETECT II: a study protocol for a prospective multicentre observational study to validate the UroMark assay for the detection of bladder cancer from urinary cells. BMC Cancer 2017;17:767. [Crossref] [PubMed]
- Pietrusiński M. Detection of bladder cancer in urine sediments by a hypermethylation panel of selected tumor suppressor genes. Cancer Biomark 2017;18:47-59. [Crossref] [PubMed]
- Bosschieter J, Bach S, Bijnsdorp IV, et al. A protocol for urine collection and storage prior to DNA methylation analysis. PLoS One 2018;13:e0200906 [Crossref] [PubMed]
- Rose M, Bringezu S, Godfrey L, et al. ITIH5 and ECRG4 DNA Methylation Biomarker Test (EI-BLA) for Urine-Based Non-Invasive Detection of Bladder Cancer. Int J Mol Sci 2020;21:1117. [Crossref] [PubMed]
- van Kessel KE, Beukers W, Lurkin I, et al. Validation of a DNA Methylation-Mutation Urine Assay to Select Patients with Hematuria for Cystoscopy. J Urol 2017;197:590-5. [Crossref] [PubMed]
- Ellinger J, El Kassem N, Heukamp LC, et al. Hypermethylation of cell-free serum DNA indicates worse outcome in patients with bladder cancer. J Urol 2008;179:346-52. [Crossref] [PubMed]
- Jabłonowski Z, Reszka E, Gromadzinska J, et al. Hypermethylation of p16 and DAPK promoter gene regions in patients with non-invasive urinary bladder cancer. Arch Med Sci 2011;7:512-6. [Crossref] [PubMed]
- Springer SU, Chen CH, Rodriguez Pena MDC, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife 2018;7:e32143 [Crossref] [PubMed]
- Hayashi Y, Fujita K, Matsuzaki K, et al. Clinical Significance of Hotspot Mutation Analysis of Urinary Cell-Free DNA in Urothelial Bladder Cancer. Front Oncol 2020;10:755. [Crossref] [PubMed]
- Christensen E, Birkenkamp-Demtroder K, Nordentoft I, et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur Urol 2017;71:961-9. [Crossref] [PubMed]
- Avogbe PH, Manel A, Vian E, et al. Urinary TERT promoter mutations as non-invasive biomarkers for the comprehensive detection of urothelial cancer. EBioMedicine 2019;44:431-8. [Crossref] [PubMed]
- Patel KM, van der Vos KE, Smith CG, et al. Association Of Plasma And Urinary Mutant DNA With Clinical Outcomes In Muscle Invasive Bladder Cancer. Sci Rep 2017;7:5554. [Crossref] [PubMed]
- Hirotsu Y, Yokoyama H, Amemiya K, et al. Genomic profile of urine has high diagnostic sensitivity compared to cytology in non-invasive urothelial bladder cancer. Cancer Sci 2019;110:3235-43. [Crossref] [PubMed]
- Birkenkamp-Demtröder K, Nordentoft I, Christensen E, et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur Urol 2016;70:75-82. [Crossref] [PubMed]
- Lee DH, Yoon H, Park S, et al. Urinary Exosomal and cell-free DNA Detects Somatic Mutation and Copy Number Alteration in Urothelial Carcinoma of Bladder. Sci Rep 2018;8:14707. [Crossref] [PubMed]
- Russo IJ, Ju Y, Gordon NS, et al. Toward Personalised Liquid Biopsies for Urothelial Carcinoma: Characterisation of ddPCR and Urinary cfDNA for the Detection of the TERT 228 G>A/T Mutation. Bladder Cancer 2018;4:41-8. [Crossref] [PubMed]
- Hayashi Y, Fujita K, Matsuzaki K, et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci 2019;110:1771-9. [Crossref] [PubMed]
- Lu H, Liang Y, Guan B, et al. Aristolochic acid mutational signature defines the low-risk subtype in upper tract urothelial carcinoma. Theranostics 2020;10:4323-33. [Crossref] [PubMed]
- Stasik S, Salomo K, Heberling U, et al. Evaluation of TERT promoter mutations in urinary cell-free DNA and sediment DNA for detection of bladder cancer. Clin Biochem 2019;64:60-3. [Crossref] [PubMed]
- Ou Z, Li K, Yang T, et al. Detection of bladder cancer using urinary cell-free DNA and cellular DNA. Clin Transl Med 2020;9:4. [Crossref] [PubMed]
- Hayashi Y, Fujita K, Nojima S, et al. TERT C228T mutation in non-malignant bladder urothelium is associated with intravesical recurrence for patients with non-muscle invasive bladder cancer. Mol Oncol 2020;14:2375-83. [Crossref] [PubMed]