
A comprehensive evaluation of differentially expressed mRNAs and lncRNAs in cystitis glandularis with gene ontology, KEGG pathway, and ceRNA network analysis
Introduction
Cystitis glandularis (CG) is a proliferative disorder of the bladder mucosa characterized by transitional cell glandular metaplasia. Typical and intestinal types of CG have been described (1). CG has been occasionally demonstrated to be a locally aggressive benign lesion (2). While it is reported that CG was associated with adenocarcinoma of the bladder, the causal relationship has yet to be fully established (3).
The pathogenesis of CG is not well understood. Through immunohistochemical techniques and in situ hybridization, expression of P16 in samples with CG were found to be abnormal, with expression in normal bladder mucosa defined as normal (4). After treatment, expression levels of p53 and Ki-67 were significantly lower in the bladder mucosa of patients with CG, suggesting that p53 may be involved in the development of CG (5). A recent comparative analysis of RNA sequencing of CG tissues and normal control tissues revealed differential expression (DE) of multiple genes including CX3CL1, CXCL6 and CXCL1, suggesting that these genes may be associated with the pathogenesis of CG (6). However, these limited studies do not effectively assess a correlation between molecular markers and the development of CG.
The understanding of the cellular control of genetic expression is rapidly expanding. Recently, gene expression has been shown to be inhibited by small, non-coding RNAs called microRNAs (miRNAs). miRNAs are RNAs of approximately 20 nucleotides in length that undergo base-pairing with complementary sequences in messenger RNA (mRNA), resulting in cleavage of the mRNA, destabilization of the mRNA, and less efficient translation of the mRNA. Individual miRNA can target many different genes. This miRNA-mediated post-transcriptional regulation of gene expression is believed to be present in half of all genes (7).
One type of long non-coding RNAs (lncRNAs) can bind to miRNAs and alter their inhibitory effect on mRNA expression, which adds another layer of control to mRNA expression (8). These lncRNAs are also called competing endogenous RNAs (ceRNAs) (9-12). The biological function of lncRNA has received great attention in recent years. Although it does not encode proteins, lncRNA does participate in many important biological processes (BP) (13-15). For example, Montes et al. reported that lncRNA MIR31HG targets p16 expression to regulate senescence (16). Additionally, Liu et al. found that lncRNA-UCA1 was regulated by p27 protein expression in primary cardiomyocytes to promote apoptosis (17).
We previously reported that several non-coding RNAs have been discovered, and we also found that lncRNA UCA1 plays an important role in the progression of CG (18-22). In this study, we aimed to investigate the role of novel lncRNAs expression in patients with CG. We used microarray screening to analyze the expression profiles of lncRNAs and mRNAs in normal and CG bladder tissues. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to predict the biological roles and potential signaling pathways of these differentially expressed lncRNAs. A coding-non-coding co-expression (CNC) network and ceRNAs analysis were performed to explore the potential roles of differentially expressed lncRNAs in CG pathogenesis.
Methods
Source of specimens
Tissue from three cases of typical CG and three normal bladders (N) were obtained from the Department of Urology, Xiangya Hospital. Diagnosis was made by pathologic examination of biopsied tissue. No patient had any other urinary disease. All specimens were stored in liquid nitrogen immediately after surgical resection and were transferred to a freezer maintained at –80 °C.
RNA extraction
Tissue was homogenized using TRIzol Reagent (Invitrogen, Grand Island, NY, USA), and total RNA was extracted in accordance with the manufacturer’s instructions. RNA quantity and quality were measured using a Nano Drop ND-1000 Spectrophotometer (ThermoFisher Scientific, Wilmington, DE, USA). RNA integrity was assessed using standard denaturing agarose gel electrophoresis.
Microarray analysis
Sample labelling and array hybridization were applied in the light of the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology, Santa Clara, CA, USA), with minor modifications. In brief, mRNA was purified from total RNA after the removal of rRNA using an mRNA-ONLYTM Eukaryotic mRNA Isolation Kit (Epicentre, Madison, WI, USA). Each sample was then amplified and transcribed along the full length of the transcript into fluorescent complimentary RNA (cRNA) without 3' bias using random priming methods (Arraystar Flash RNA Labelling Kit, Arraystar, Rockville, MD, USA). The labeled cRNAs were purified using a RNeasy Mini Kit (Qiagen, Hilden, Germany). The concentration (µg cRNA) and specific activity (pmol Cy3/µg cRNA) of the labeled cRNAs were determined using a NanoDrop ND-1000 Spectrophotometer.
Labeled cRNA from each tissue specimen was fragmented according to a modified protocol. In brief, cRNA was placed in 10× blocking agent and 25× fragmentation buffer and incubated at 60 °C for 30 min. Then 2× GE hybridization buffer was added to dilute the labeled cRNA to stop the fragmentation reaction. Subsequently, hybridization solution was dispensed into the gasket slide, which was then assembled with the lncRNA expression microarray slide. The slides were incubated for 17 h at 65 °C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed and scanned using an Agilent Technologies G2505C SureScan High-Resolution DNA Microarray Scanner (Agilent Technologies, Santa Clara, CA, USA). Microarray hybridization and expression data collection were performed by KangChen Bio-tech, Shanghai, China.
A two-fold change (FC) in expression between CG and normal tissues was considered significant. This was calculated as log2 (CG/N). All values greater than 0.58 were up-regulated and all values less than –0.58 were considered down-regulated genes.
Quantitative real-time PCR (qRT-PCR) validation
Total RNA was isolated from tissues to generate cDNA using the SuperScript III Reverse Transcriptase (Invitrogen, Grand Island, NY, USA). Each cDNA sample was amplified using a Biosystems ViiA 7 Real-Time PCR System. Each cycle consisted of denaturation at 95 °C for 10 s and extension at 60 °C for 60 s. β-actin was used as an endogenous control to normalize each sample. The primers are listed in Table 1.
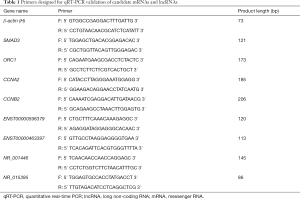
Full table
GO and KEGG pathways analysis
The GO project provides a controlled vocabulary to describe gene and gene product attributes in any organism (http://www.geneontology.org). The ontology covers three domains: BP, cellular components (CC) and molecular functions (MF). A Fisher’s exact test was used to the compare expression of CG and normal tissues of items found in the DE and GO annotation lists. P values less than 0.05 were considered statistically significant. KEGG pathways (http://www.genome.jp/kegg/) were used to describe molecular interactions, reactions, and relation networks of identified genes and gene products.
CNC network analysis
To identify interactions among the differentially expressed mRNAs and lncRNAs, we constructed a CNC network. The network was constructed according to the normalized signal intensities of specific mRNA and lncRNA expression levels. A Pearson correlation coefficient >0.95 was used as a cutoff to identify coding and non-coding genes.
ceRNA network analysis
lncRNAs and mRNAs from CG specimens with significantly different expression than that found in normal tissues were subjected to ceRNA network nalysis in order to identify potential miRNA response elements (MREs). Overlapping of the same miRNA seed sequence binding site on both lncRNAs and mRNA sequences was identified in order to predict potential interactions. Potential targets of miRNAs were predicted using our miRNA target prediction software that is based on TargetScan (http://www.targetscan.org/) & miRanda (http://www.mircorna.org/).
Results
Microarray analysis
Microarray analysis identified 436 significantly dysregulated lncRNAs in CG tissues. Among them, 198 were up-regulated and 238 were down-regulated (greater than 2-fold difference as compared to normal tissue, P<0.05). The top 20 most significantly dysregulated lncRNAs are listed in Table 2.
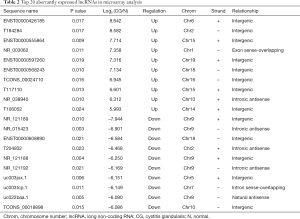
Full table
A total of 809 significantly dysregulated mRNAs were identified in CG tissues. Among them, 606 were up-regulated and 203 were down-regulated (greater than 2-fold difference as compared to normal tissue, P<0.05). The top 20 most significantly dysregulated mRNAs are listed in Table 3.
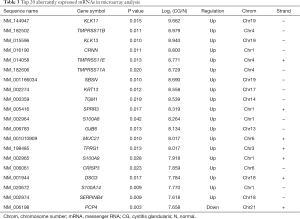
Full table
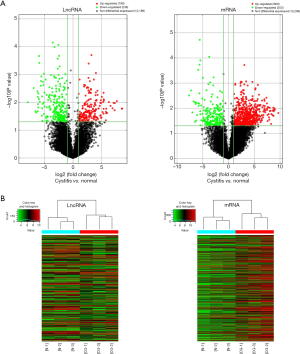
The variation of lncRNA and mRNA expression between CG and normal tissues is displayed in a Volcano plot (Figure 1A). Cluster analysis was performed to demonstrate relationships among lncRNA and mRNA expression patterns in normal bladder mucosa and CG (Figure 1B).
GO and KEGG pathway analysis
Over- and under-expressed mRNAs that co-expressed with each of the differentially expressed lncRNAs were identified and listed as genes or gene products in the GO project (Figure 2A). The most enriched GO terms were epidermis development (ontology: BP, GO: 0008544), extracellular exosome (ontology: CC, GO: 0070062) and protein binding involved in cell-cell adhesion (ontology: MF, GO: 0098632). The most highly enriched GO sequences for the down-regulated transcripts were muscle system process (ontology: BP, GO: 0003012), contractile fiber part (ontology: CC, GO: 0044449) and carbohydrate derivative binding (ontology: MF, GO: 0097367).
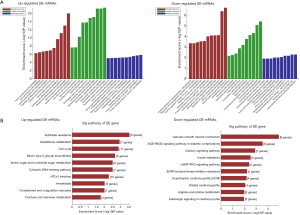
KEGG pathway analysis indicated that the mRNAs co-expressed with lncRNAs were involved in the regulation of cell cycle. The top 10 KEGG pathways are listed in Figure 2B.
CNC network analysis
Sequences that affect cell cycle progression were found in microarray analyses to be dysregulated in CG. Four related mRNAs, mothers against decapentaplegic homolog 3 (SMAD3), origin recognition complex subunit 1 (ORC1), cyclin A2 (CCNA2), and cyclin B2 (CCNB2), were selected for verification by qRT-PCR (Figure 3A).

Pearson coefficient analysis revealed that expression of these four genes was associated with expression of 37 other genes. Co-expression network profile analysis demonstrated 41 network nodes and 71 connections among the four mRNAs and 37 differentially expressed lncRNAs (Figure 3B). There were 24 negative and 47 positive correlations within the network. Four up-regulated lncRNAs were randomly chosen for qRT-PCR validation (ENST00000596379, ENST00000463397, NR_001446 and NR_015395). As expected, the four lncRNAs were up-regulated in the CG tissues compared to normal tissues (greater than 2-fold, P<0.05) (Figure 3C), which is consistent with the microarray chip data.
ceRNA network
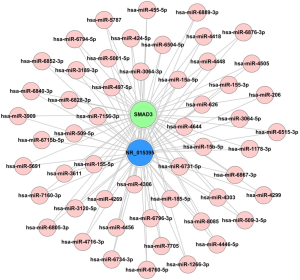
Different ceRNAs have been shown to compete for the same MREs, regulating different levels of expression for each ceRNA (8). To investigate whether the four lncRNAs (ENST00000596379, ENST00000463397, NR_001446 and NR_015395) and four mRNAs (SMAD3, ORC1, CCNA2 and CCNB2) share any common MRE binding sites, a ceRNA network in CG was constructed with these elements from the microarray data. NR_015395 is a ceRNA of miR-133a-3p that targets SMAD3. Pearson coefficient analysis showed that expression of these two elements was correlated with the expression of 53 other genes (Figure 4).
Discussion
Cellular proliferation and glandular metaplasia were observed in the transformation of normal bladder mucosa to CG. These changes were caused by alterations in the regulatory mechanisms that control normal cellular maintenance and growth-related functions. We examined the expression of mRNAs and lncRNAs in normal bladder mucosa and CG in order to identify abnormally expressed genetic material associated with the development of CG.
Using a second-generation lncRNA microarray, 809 mRNAs with abnormal expression in CG were identified. Of these, 606 mRNAs were found to be significantly up-regulated in CG as compared to normal bladder tissue (FC ≥2.0, P<0.05). Four mRNAs were related to cell cycle progression, SMAD3, ORC1, CCNA2 and CCNB2, and had increased mRNA expression in CG. This increased expression was verified with qRT-PCR.
Several studies have shown that lncRNAs play a significant role in regulating cell cycle activity (23-27). Microarray analysis identified 436 lncRNAs abnormally expressed in CG. Of these, 198 were found to be significantly up-regulated in CG (FC ≥2.0, P<0.05). GO and KEGG pathway analyses were performed to predict the potential functions of differentially expressed lncRNAs. GO analysis identified lncRNAs involved in epidermis development, extracellular exosome and protein binding related to cell-cell adhesion. KEGG pathway analysis indicated that the dysregulated lncRNAs were significantly associated with altered expression of cell cycle pathways in CG.
A CNC network analysis of SMAD3, ORC1, CCNA2 and CCNB2 mRNA expression identified 37 differentially expressed lncRNAs associated with their expression. SMAD3 expression was correlated with 15 lncRNAs expression, ORC1 expression was correlated with 18 lncRNAs expression, CCNA2 expression was correlated with 15 lncRNAs expression, and CCNB2 expression was correlated with 23 lncRNAs expression. Four up-regulated lncRNAs (ENST00000596379, ENST00000463397, NR_001446 and NR_015395) that were identified in the CNC network analysis as being associated with the expression of these four mRNAs were selected randomly for qRT-PCR verification. All four lncRNAs were also significantly up-regulated in CG.
We suggest a ceRNA mechanism maybe involved in this binding because of the consistent expression trends, and there may also be some shared MRE sites. The results of ceRNA network for CG showed that NR_015395 was a ceRNA of miR-133a-3p that targets SMAD3. These findings suggest that altered lncRNA expression (ceRNA) may be involved in CG-associated signaling pathways. In future work, we plan to establish more molecular mechanism at the clinical, cellular, animal, and molecular levels to provide new experimental evidence for the pathogenesis of CG. Solving these key problems will help us understand the molecular mechanism underlying the occurrence and development of CG and provide a theoretical basis as well as drug targets for new prevention and treatment strategies to inhibit dysuria through interruption of the inflammatory response.
This study has some limitations. A small number of tissue samples were examined. The analysis method identifies first-order miRNA-mediated lncRNA-mRNA associations but does not identify other influences. This method does not verify targets regulated by translational repression because it is based on changes in the transcript level. Further validation of the identified associations is needed.
Conclusions
Collectively, microarray data was used to identify mRNAs, lncRNAs, and ceRNAs with significantly altered expression profiles in CG. These findings were validated using qRT-PCR. Dysregulated mRNA from BP, CC and MF were unveiled. A significant dysregulation of lncRNAs was also observed. While these findings provide novel insights to the latent role of lncRNAs in CG, further research is required to fully explain their role in inducing transitional cell glandular metaplasia.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding: This work was supported by National Nature Science Foundation of the People’s Republic of China (81873626 and 81700665).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.03.01). XG and XZ serves as an unpaid editorial board member of Translational Andrology and Urology from Mar 2019 to Feb 2021. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Review Board of Xiangya Hospital, Central South University (Changsha, China). Informed consent was obtained from all patients (ethical approval ID: 201803086).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung MT, Lopez-Beltran A, Eble JN, et al. Divergent pathway of intestinal metaplasia and cystitis glandularis of the urinary bladder. Mod Pathol 2006;19:1395-401. [Crossref] [PubMed]
- Zhu JX, Gabril MY, Sener A. A rare case of recurrent urinary obstruction and acute renal failure from cystitis cystica et glandularis. Can Urol Assoc J 2012;6:E72-4. [Crossref] [PubMed]
- Behzatoğlu K. Malignant glandular lesions and glandular differentiation in invasive/noninvasive urothelial carcinoma of the urinary bladder. Ann Diagn Pathol 2011;15:422-6. [Crossref] [PubMed]
- Schwartz LE, Khani F, Bishop JA, et al. Carcinoma of the uterine cervix involving the genitourinary tract: a potential diagnostic dilemma. Am J Surg Pathol 2016;40:27-35. [Crossref] [PubMed]
- Li A, Zhou J, Lu H, et al. Pathological feature and immunoprofile of cystitis glandularis accompanied with upper urinary tract obstruction. Biomed Res Int 2014;2014:872170. [PubMed]
- Chu CL, Zhao CH, Zhang ZW, et al. Identification and validation of gene expression patterns in cystitis glandularis patients and controls. SLAS Discov 2017;22:743-50. [Crossref] [PubMed]
- Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 2002;30:363-4. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Liao C, Long Z, Zhang X, et al. LncARSR sponges miR-129-5p to promote proliferation and metastasis of bladder cancer cells through increasing SOX4 expression. Int J Biol Sci 2020;16:1-11. [Crossref] [PubMed]
- Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer 2017;16:136. [Crossref] [PubMed]
- Wang J, Zhang H, Situ J, et al. KCNQ1OT1 aggravates cell proliferation and migration in bladder cancer through modulating miR-145-5p/PCBP2 axis. Cancer Cell Int 2019;19:325. [Crossref] [PubMed]
- Li MJ, Zhang J, Liang Q, et al. Exploring genetic associations with ceRNA regulation in the human genome. Nucleic Acids Res 2017;45:5653-65. [Crossref] [PubMed]
- Seo JS, Diloknawarit P, Park BS, et al. ELF18-INDUCED LONG NONCODING RNA 1 evicts fibrillarin from mediator subunit to enhance PATHOGENESIS-RELATED GENE 1 (PR1) expression. New Phytol 2019;221:2067-79. [Crossref] [PubMed]
- Xu M, Xiang Y, Liu X, et al. Long noncoding RNA SMRG regulates Drosophila macrochaetes by antagonizing scute through E(spl)mβ. RNA Biol 2019;16:42-53. [Crossref] [PubMed]
- Wang J, Chen X, Shen D, et al. A long noncoding RNA NR_045363 controls cardiomyocyte proliferation and cardiac repair. J Mol Cell Cardiol 2019;127:105-14. [Crossref] [PubMed]
- Montes M, Nielsen MM, Maglieri G, et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun 2015;6:6967. [Crossref] [PubMed]
- Liu Y, Zhou D, Li G, et al. Long non coding RNA-UCA1 contributes to cardiomyocyte apoptosis by suppression of p27 expression. Cell Physiol Biochem 2015;35:1986-98. [Crossref] [PubMed]
- Li C, Cui Y, Liu LF, et al. High expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin Genitourin Cancer 2017;15:570-6. [Crossref] [PubMed]
- Li Q, Li C, Chen J, et al. High expression of long noncoding RNA NORAD indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Urol Oncol 2018;36:310.e15-22. [Crossref] [PubMed]
- Chen JB, Zhu YW, Guo X, et al. Microarray expression profiles analysis revealed lncRNA OXCT1-AS1 promoted bladder cancer cell aggressiveness via miR-455-5p/JAK1 signaling. J Cell Physiol 2019;234:13592-601. [Crossref] [PubMed]
- Yu C, Longfei L, Long W, et al. LncRNA PVT1 regulates VEGFC through inhibiting miR-128 in bladder cancer cells. J Cell Physiol 2019;234:1346-53. [Crossref] [PubMed]
- Zhou X, Cui Y, Chen J, et al. UCA1 promotes cell viability, proliferation and migration potential through UCA1/miR-204/CCND2 pathway in primary cystitis glandularis cells. Biomed Pharmacother 2019;114:108872. [Crossref] [PubMed]
- Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 2018;33:706-20.e9. [Crossref] [PubMed]
- Liu Z, Chen Z, Fan R, et al. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer 2017;16:82. [Crossref] [PubMed]
- Liu M, Zhang H, Li Y, et al. HOTAIR, a long noncoding RNA, is a marker of abnormal cell cycle regulation in lung cancer. Cancer Sci 2018;109:2717-33. [Crossref] [PubMed]
- Jin F, Wang N, Zhu Y, et al. Downregulation of long noncoding RNA Gas5 affects cell cycle and insulin secretion in mouse pancreatic β cells. Cell Physiol Biochem 2017;43:2062-73. [Crossref] [PubMed]
- Hu YW, Kang CM, Zhao JJ, et al. LncRNA PLAC2 down-regulates RPL36 expression and blocks cell cycle progression in glioma through a mechanism involving STAT1. J Cell Mol Med 2018;22:497-510. [Crossref] [PubMed]


