
Future diagnostics in male infertility: genomics, epigenetics, metabolomics and proteomics
Introduction
Infertility is defined as inability to conceive within 12 months of regular unprotected sexual intercourse (1) and affects 15% of couples (2). A male factor alone contributes to 30% of these cases, and to another 20% of cases when combined with female factors (3). In addition to the medical history and physical exam, the workup of male factor infertility centers on the endocrine evaluation and semen analysis (SA). Though the SA remains a mainstay of the infertility workup, it is an imperfect test with significant variability and only modest predictive value for fertility (4). While additional serum blood tests such as testosterone, luteinizing hormone, follicle stimulating hormone and prolactin may point the clinician towards an underlying etiology for male factor infertility, these tests in combination with the SA still may not provide a diagnosis. This has led to the need to develop adjunct tests to assist with diagnosis of the underlying etiology of infertility and subfertility which may be reflected in an abnormal SA (5).
Genetics have long been recognized as playing a role in male factor infertility. The most common genetic causes include azoospermia factor (AZF) deletions of the Y chromosome, Klinefelter syndrome with an extra copy of the X chromosome, and monogenic disorders including Kallman syndrome and cystic fibrosis. However, these genetic conditions account for only 30% of cases of male infertility and subfertility (6,7), with approximately 50% of male factor infertility still classified as idiopathic, after controlling for hormonal abnormalities or other anatomic etiologies (6). The proportion of these cases that are due to genetic causes remains unknown (7). Spermatogenesis is a complex process involving the coordination of hormones, environmental factors, and over 2,000 different genes (8). Animal models have demonstrated that the knockout of numerous genes leads to subfertility (9) and the desire to identify an etiology of infertility has driven investigation into identification of the genes involved, epigenetic changes to these genes, the role of these changes on subfertility and infertility, and identification of biomarkers which may improve the diagnostic capability of semen parameters.
The aim of this review is to outline the role of biomarkers in the diagnosis of male infertility, and to define the current role and future use of genomics, epigenetics, metabolomics and proteomics in the workup of the infertile or subfertile male.
Biomarkers in male infertility
Biomarkers
A biomarker is defined as a quantifiable and measurable biological parameter which serves as an index for certain disease states (10). The ideal biomarker would be cost-effective, minimally invasive, accurate, and able to detect disease in the early stage. In the field of infertility, the SA acts as a basic biomarker for male infertility. However, the SA remains an imperfect test as it may be affected by physiologic parameters including abstinence interval, systemic illness, medical comorbidities, life style factors including diet and activity, and environmental exposures (11-17). Given the high variability in semen parameters, multiple adjunct biomarkers have been developed over the years, including antisperm antibody (ASA) testing, acrosome reaction testing, sperm penetration assays, sperm-zona pellucida binding tests, hyaluronan binding assays, and deoxyribonucleic acid (DNA) damage testing (18-21).
ASA testing was one of the first widely-used biomarkers in the field of male infertility. ASAs develop with disruption of the blood-testis barrier as in the case of trauma, infection, testicular torsion, or even following vasectomy (22). ASAs have been identified in approximately 12% of infertile men, versus 2.5% of fertile men (22). While ASAs are thought to have a negative impact on fertility by impacting the quality of sperm, many studies on the effect of ASA on semen parameters have shown conflicting results (18). A recent meta-analysis by Cui et al. suggested that ASA may have a negative effect on sperm concentration and progressive motility (18). However, other studies have shown that the presence of ASAs does not translate into consistently altered rates of spontaneous, intrauterine insemination (IUI)-assisted, in vitro fertilization (IVF)-assisted, or intracytoplasmic sperm injection (ICSI)-assisted pregnancies (23-25), which brings into question the utility of ASA as a biomarker for male infertility.
Numerous sperm function tests have been developed, including assays for detecting the acrosome reaction, sperm penetration, sperm-zona pellucida binding, and hyaluronan binding. In the era of ICSI, functional sperm tests have fallen out of favor as the functional aspects of the sperm can be bypassed with direct intracytoplasmic injection. However, these tests retain some utility as they may allow the provider to direct a patient away from futile rounds of IUI or IVF in certain situations (19). Sperm function tests have been shown to be highly predictive of the sperm’s ability to fertilize an egg, with the downside of these tests being the requirement for intact sperm or eggs, as well as the time consuming and technically challenging nature of these tests, thus limiting their utility as a biomarker (19).
DNA fragmentation has been associated with impairments in natural conception, IUI, and IVF (3,26,27), with inconsistent effects on outcomes with ICSI (28). Recent guidelines suggest performing DNA fragmentation screening on men with a clinical varicocele and borderline to normal semen parameters to help identify optimal surgical candidates, men with unexplained infertility, recurrent spontaneous pregnancy loss, recurrent IUI, IVF or ICSI failure, or patients with lifestyle risk factors including smoking, advanced age, obesity, or environmental exposures (29). Based on these guidelines, O’Neill et al. proposed a treatment algorithm for patients with poor IUI outcomes and suggested that these patients undergo DNA fragmentation testing (28). Patients with a normal DNA fragmentation assay proceeded to IVF, and patients with an abnormal assay proceeded to ICSI. If subsequent IVF or ICSI was unsuccessful, patients were encouraged to pursue epididymal or testicular sperm extraction, as studies have shown that levels of DNA damage are higher in ejaculated sperm (30). This group suggested that this algorithm could be used to guide the decision regarding the assisted reproductive technology (ART) treatment approach, in hopes of decreasing the number of futile treatment cycles and shorten the time to conception (28). When determining the utility of proceeding with multiple rounds of ART versus preceding directly to testicular sperm extraction for patients with high DNA fragmentation, the provider must utilize shared decision making with the patient and the partner, and fully consider the costs of multiple rounds of ART and risks incurred by the female partner weighed against the risks to the male patient undergoing extraction. A recent review of the literature by Awaga et al. showed that the use of testicular spermatozoa, rather than ejaculated spermatozoa, results in a higher probability of pregnancy in patients with high DNA fragmentation index, but only in the setting of oligospermia (31). Given this, it may be beneficial to proceed with sperm extraction and ICSI for men found to have high levels of DNA fragmentation in the setting of oligospermia, rather than attempting ICSI with ejaculated spermatozoa.
The limited utility of the historically used biomarkers in identifying the underlying etiology of male infertility has prompted the search for markers which would be easier to employ, more cost effective, and more predictive of reproductive outcomes. Since the mapping of the human genome, much interest has been paid to the field of genetic medicine, with treatment modalities tailored to a patient’s specific genetic makeup. The study of genomics, including epigenetics, metabolomics, and proteomics, holds promise as a potential area of targeted diagnostics in the setting of male infertility. The following sections will review the recent advances in, and future directions of, genomics and its sub-fields.
Genomics
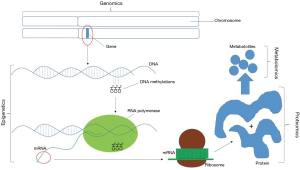
The field of genomics centers on the systematic evaluation of an organism’s complete DNA sequence (Figure 1) (32). Since the advent of karyotyping, the hunt for genetic causes of infertility has gained interest. Following the initial cloning of the Y chromosome in 1992, the first genetic marker of infertility, AZF, was identified (33-35). It was soon determined that the presence of specific sub-regions of AZF is critical for proper spermatogenesis. Deletion of the AZFa region leads to deletion of the USP9Y and DBY/DDX3Y genes resulting in Sertoli cell only syndrome. Deletion of the AZFb region results in deletion of the RBMY1 and PRY genes which causes arrest of spermatogenesis. Though the AZFc region also contains genes important in spermatogenesis, microdeletions in this region typically allow for limited spermatogenesis, therefore making these patients candidates for testicular sperm extraction (7,36). These microdeletions are typically assessed using polymerase chain reaction (PCR).
Another common application of the field of genomics as it relates to male infertility is in the workup of congenital bilateral absence of the vas deferens (CBAVD). CBAVD occurs in approximately 1% of infertile men, is due to mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, and results in obstructive azoospermia (7). Standard CFTR screening via PCR is recommended for patients presenting with CBAVD. In cases where standard CFTR screening is negative, and the female partner is a carrier, full gene sequencing may be considered (37).
In its most basic form, genomics includes standard karyotyping. It has been reported that chromosomal abnormalities are 8–10 times more common in infertile men, with anomalies occurring in approximately 3% of patients with oligospermia, and 19% of patients with azoospermia (7,38). The karyotype is one of the most common tests recommended for the workup of male infertility and is recommended by the American Urological Association (AUA) in patients with severe oligospermia of <5 million sperm/mL and non-obstructive azoospermia (37). However, studies have suggested that chromosomal abnormalities are identified in patients with less severe oligospermia, and that this stringent cutoff may fail to capture a significant number or patients with chromosomal abnormalities in the setting of unexplained infertility (39). A recent study of over 16,000 men showed that the frequency of major chromosomal anomalies in men with primary infertility was 5.5%, and 2.5% of men with secondary infertility, with fewer abnormalities noted in patients with normal semen analyses (SA) (40). Patients with a history of adverse pregnancy outcomes were found to have chromosomal abnormalities in 1.8% of cases regardless of SA results. Out of all patients evaluated, 4% were noted to have chromosomal abnormalities. 64% of these cases involved changes in autosomal chromosome number or structure, and 31% involved changes in sex chromosome number or structure. Even in patients with a normal SA, chromosomal abnormalities were still detected in 0.02% of patients with primary infertility and 0.3% of patients with secondary infertility, suggesting that chromosomal anomalies may contribute to infertility even in the presence of a normal SA (40). In addition to well-described abnormalities in sex chromosomes in patients with infertility, this group found a high occurrence of chromosomal polymorphic variations. These variations have been shown to be associated with poor spermatogenesis and infertility, though the exact mechanism of these remains unclear (40).
While cytogenetic karyotyping of peripheral blood lymphocytes can provide basic information regarding overall chromosome number and structure, fluorescent in situ hybridization (FISH) is gaining popularity in the field of genomics to identify specific chromosomal regions which may be associated with deleterious effects on semen quality and fertility. FISH has been used since the 1990s to assess sperm for aneuploidy of chromosomes X, Y, 13, 18 and 21, which are thought to be most frequently involved in spontaneous miscarriage (41). The downfall of FISH analysis on sperm is the high cost associated with the testing, and the fact that the sperm which are tested are not able to be utilized for ART. However, knowledge of the presence of spermatic aneuploidy may help in counseling of patients regarding the risks of ICSI and whether to proceed with preimplantation genetic testing (3).
Mayeur et al. have suggested that a criterion of <5×106 progressive spermatozoa retrieved after sperm preparation may be a better threshold for genetic screening in patients with infertility, to better capture patients with Robertsonian and reciprocal translocations which may be playing a role in infertility (39). Their studies, along with others, have identified numerous chromosomes (mostly commonly chromosomes 1, 2, 6, 9, 16, and 17) with redundant regions or polymorphisms which were associated with infertility and poor or very poor progressive spermatozoa after processing. This suggests that these regions may contain genes which are important for proper sperm function (39,40).
As additional genes are being identified as contributing to male infertility, FISH may begin to play a bigger role in the assessment of peripheral lymphocytes for Robertsonian translocations and reciprocal translocations which may impact fertility, without having to test, and thus destroy, viable sperm which may be present in small numbers and utilized in ART. One challenge to using FISH as a diagnostic tool, however, is that the DNA sequence of interest must be known prior to the application of the fluorescent label. It should be recognized that due to the complex interplay of the thousands of genes involved, the phenotype of infertility or subfertility may not be uniform, which makes the search for the genomics involved more difficult (7).
Thousands of genes are involved in spermatogenesis. With the advent of whole-genome assessments using comparative genomic hybridization (CGH), high-throughput sequencing technologies or next generation sequencing, and analysis of single nucleotide polymorphism (SNP) arrays, it is becoming possible to study large amounts of genetic material for relatively small sequences of abnormalities which may impact fertility. These technologies have shown promise in terms of effectiveness and affordability in identifying potential biomarkers and candidate genes in male infertility. An excellent review by Krausz and Riera-Escamilla detailed these techniques, described their utility, and several of the genes implicated in male infertility (42). Okutman et al. have recently compiled a list of 31 genes for which the data is strong to support their role in abnormal spermatogenesis or fertilization failure (43). Though their study was published in November of 2018, Cannarella et al. have even more recently published a list of 60 genes involved in spermatogenic failure (44), which speaks to the exponential expansion of our knowledge of the genes involved in male infertility. Similarly, Patel et al. recently demonstrated the utility of using next generation sequencing to identify genes known to impact male fertility, suggesting that next generation sequencing may be used to more rapidly identify a larger number of genes thought to be related to male infertility with greater accuracy and at significantly reduced cost compared to traditional standard analysis (45). However, though we continue to identify genes involved in infertility, these genes remain primarily of diagnostic interest, and few have resulted in therapeutic targets for the treatment of male infertility. As we gain a deeper understanding of the genes involved, karyotyping, FISH, and gene sequencing may be able to further stratify male infertility into phenotypic subsets.
Epigenetics:
Genetic epigenesis is the process of modifying the DNA of a cell to ultimately impact cell differentiation. Epigenetic changes to DNA, including DNA methylation, heterochromatization, histone modification, timing of DNA replication, and nucleosome positioning cause heritable changes to cells without altering the DNA sequence (Figure 1) (46). This process is well known to impact gene expression in somatic ells, and it is being shown to have a role in the sperm as well. Spermatozoa are particularly susceptible to oxidative stress, and reactive oxygen species (ROS) can induce epigenetic changes in sperm DNA (47). It is thought that sperm histone modifications, DNA methylation, and protamination may impact transcription during embryogenesis (48), and some evidence suggests that these changes could result in miscarriage (49).
The use of epigenetic changes in DNA as a marker to identify male factor infertility is difficult, as these changes may have a subtle or even insignificant biological impact, and multiple different changes may lead to similar infertility phenotypes. While determining causal relationships between these changes and embryologic outcomes remains challenging, several spermatic epigenetic changes have been associated with sperm abnormalities. For example, individual sperm with low levels of protamine-1 and protamine-2 content have been shown to have increased susceptibility to DNA damage and decreased viability (50). Spermatic DNA hypomethylation of the gene H19, and hypermethylation of genes MEST and SRNPN have been associated male infertility (51). Additionally, alterations in DNA methylation at specific genomic loci have been associated with difficulty conceiving (52), and DNA methylation and gene transcription patterns may be predictive of success rates of IUI and IVF (5,53). It has also been shown that patients with a history of oligoasthenoteratozoospermia (OAT) have higher rates of DNA methylations and altered gene expression in embryos that have undergone preimplantation genetic screening following IVF (49). DNA methylation as a marker of male infertility is an attractive target, as DNA methylation remains stable throughout spermatogenesis, unlike RNA transcription and histone modification (48). Whether epigenetic changes in sperm represent causative etiologies of infertility and subfertility, or whether these changes reflect an alternate underlying pathology, remains to be determined.
With the growing concern about the decline in male fertility (54), much research is being targeted towards the impact of environmental factors on sperm quality. It is known that oxidative stress and urinary phthalates and phthalate alternatives impair sperm DNA methylation (55). There is concern that environmental factors, particularly plastic-derived compounds, may generate oxidative stress leading to heritable abnormal DNA methylation (56). In addition to ingested environmental contaminants, the increase in rates of obesity and diabetes have been associated with male infertility and decreased sperm quality (55,57). Though this is related in part to obesity-induced endocrine disruption, obesity is thought to also result in epigenetic changes in sperm, the impacts of which may lead to health impairments in offspring (58). Currently, the only commercially available screening tool for epigenetic changes in sperm is “Seed” developed by Episona Inc., developed in 2016.With the growing interest in epigenetics as a marker for male infertility, it is expected that additional companies will develop testing for these factors.
Post-transcriptional epigenetics may also play a role in male infertility. This is largely performed by short non-coding ribonucleic acids (sRNAs), including microRNAs, which modulate protein translation (59). These sRNAs are expressed in germ cells and are required for normal spermatogenesis, and numerous microRNAs have been found to be up- or down-regulated in the sperm and seminal plasma of men with subfertility and infertility (59,60). Additional modifications to the sperm epigenome take place in the epididymis, as the spermatozoa undergo further maturation. Modification of sperm sRNAs during maturation in the epididymis has been linked to normal fertilization potential, embryonal development and offspring phenotype (61). It has been hypothesized that the sRNA profile of spermatozoa is altered by interaction with epididymosomes, membrane-bound extracellular vesicles which encapsulate complexes of sRNAs and proteins and affect the luminal contents of the epididymis (61). Multiple environmental stressors have been shown to alter the sRNA profile of sperm, and it is thought that this occurs through packaging of altered sRNAs under stressful conditions into epididymosomes, effecting which sRNAs are delivered to maturing sperm (61). This is of clinical interest, as it suggests that using testicular-derived sperm for embryo fertilization may bypass the negative downstream effects that altered sRNAs may have on normal fertilization and embryo phenotype.
Proteomics
Proteomics involves the study of the structure and function of the expressed complement of proteins (Figure 1) (62). This concept is important, as gene expression does not necessarily translate into protein expression, and the protein complement varies from cell to cell. This makes proteomic analysis an attractive target for biomarker development. However, the protein complement of the ejaculate is complex and is added to by the epididymis, seminal vesicles, prostate and bulbourethral glands (63). A systematic review by Amaral et al. in 2014 identified over 6,000 different sperm proteins involved in multiple functional pathways (64), and the location of these proteins on the sperm have been linked to their site of origin in the testis or epididymis (65). With the development of mass spectrometry systems, the sensitivity and specificity of, as well as the ease of, protein identification has improved (65). These techniques can identify proteins within the seminal plasma, sperm membrane, various sperm regions, or mitochondria, or perhaps more importantly protein-protein interaction networks (65). It is important to understand the individual proteins involved in various fertility processes, as well as the way these proteins interact with each other, as alterations in individual proteins may alter the downstream effect of their function and interaction, resulting in various phenotypes of infertility. Many studies have been conducted and have identified a wide array of protein complement patterns present in the setting of recurrent IVF failures, asthenozoospermia, OAT, NOA, globozoospermia, teratospermia, elevated levels of DNA fragmentation and ROS, and clinical varicoceles (66-73).
The utility of analyzing the semen proteome as a biomarker for infertility is complicated, as semen specimens are highly variable within individuals and are influenced by environmental factors, as well as post-ejaculation physiological changes to sperm (74-76). In a model of mice exposed to endocrine disruptor chemicals including compounds found in plastics, numerous proteins were found to be up- or down-regulated in the testis, the majority of which were involved in cell death (77). Additionally, it has been difficult to find proteins and peptides in sperm and semen which are reproducibly altered in various phenotypes of infertility (78). Barrachina et al. recently described a novel strategy of analysis of quantitative proteomic data based on the establishment of stable-protein pairs, which allowed for identification of highly stable proteome signatures in men with normal semen parameters. However, when they applied this analysis technique to infertile men classified according to abnormal semen parameters, a high level of heterogeneity was identified, suggesting the current classification system may not be adequate to describe various infertility phenotypes (78).
A review of the literature published as recently as January of 2019 resulted in numerous articles in the field of proteomics, demonstrating the growing interest in proteomics in the study of male infertility. One recent study comparing the proteome of the ejaculate from fertile men with high ROS levels to that of fertile men with normal ROS levels found that numerous antioxidant proteins were present in higher levels in men with elevated ROS levels, suggesting that there are specific proteins which may be upregulated in fertile men with high ROS levels acting in a protective fashion to mitigate the negative effects of ROS and preserve fertility (79). Additionally, recently heat shock protein A4L has found to be downregulated in sperm in patients with asthenozoospermia, resulting in decreased sperm motility and sperm-oocyte penetration (80). This is just a small sample of the number of studies with promising results targeting specific proteins which appear to be located in the sperm and seminal plasma which may be developed as biomarkers for male infertility in the future.
Metabolomics
Metabolomics is the study of cellular metabolic products (Figure 1) (81). As metabolic products are the downstream products of gene expression, they may be a closer representation of an actual phenotype than genomics, proteomics or epigenetics (82). As previously discussed, oxidative stress can induce epigenetic changes in sperm, and leads to formation of ROS. Elevated ROS levels are seen in infertile men, and are associated with impaired sperm morphology, concentration, and motility, as well as increased levels of DNA fragmentation (3,83). ROS have gained interest as potential biomarkers of male infertility, and the study of ROS in semen is likely the most widely known application of metabolomics in male infertility. Serum samples have also been assessed for metabolomic alterations to identify a pattern in patients with NOA (84).
Interestingly, a recent study by Gilany et al. used Raman spectroscopy to identify ROS in the ejaculate of 20 patients with non-obstructive azoospermia (NOA) to detect spermatogenesis. Their assay was able to distinguish patients who had spermatogenesis on testicular sperm extraction (TESE) from those without spermatogenesis and was further able to sub-classify patients with a negative TESE into patients with hypospermatogenesis, sperm maturation arrest, and Sertoli-only syndrome (85). Another study by Gilany et al. analyzed seminal plasma of men with NOA who underwent subsequent TESE (86). Gas chromatography-mass spectrometry (GC-MS) was used to perform untargeted metabolomics profiling, and this approach was able to distinguish a differential metabolomic profile between NOA men who had sperm identified on TESE, and those with no sperm identified on TESE (86). If these results are determined to be reproducible, this technology could impact the pre-operative screening and counseling of patients with NOA.
Other metabolic products of spermatogenesis are being investigated as biomarkers for male infertility. A study by Zhao et al. utilized GC-MS to identify sperm metabolites associated with idiopathic asthenozoospermia. They identified numerous metabolites which were either upregulated or downregulated, involved in amino acid, energy, and nucleoside metabolism (87). Qiao et al. also analyzed seminal plasma using GC-MS and identified 44 metabolites that were expressed in altered amounts in patients with unexplained infertility with normal SA as compared to fertile controls, predominantly involving alterations in amino acid metabolism (88). Their metabolite profiling platform distinguished 82% of infertile patients from fertile controls and based on these results they suggested that their profiling platform may be used for diagnosis of idiopathic infertility (88). Similarly, a recent meta-analysis by Guo et al. identified 16 metabolites in seminal plasma which were significantly different between men with normal BMI and normal semen parameters versus obese men with abnormal semen parameters (89). Specifically, the pathways of arginine, proline, beta-alanine and glutathione metabolism were significantly altered, with significantly higher concentrations of spermidine and spermine identified in obese men with abnormal semen parameters (89). Spermidine and spermine have been shown to be important in normal fertility (90). Overall, metabolomic profiling of the ejaculate is a rapidly growing field, and shows promise for the development of accurate biomarkers for infertility phenotypes.
While analysis of the proteomic and metabolomic profiles of sperm and semen remain an attractive minimally invasive way to diagnose male infertility with the potential to distinguish infertility phenotypes, clinical application of these profile panels remains limited, in part due to the wide range of proteins, peptides, and metabolites found in semen samples which may be affected by various environmental factors to varying degrees between individuals. Significantly more research must be done before these profile panels can implemented clinically on a large scale.
Conclusions
Male infertility is a complex disease involving the interplay of thousands of genes, proteins, and metabolites. The study of these factors, along with epigenetic changes to the sperm DNA, remains an attractive target for the development of specific biomarkers which may be able to identify the etiology of male infertility in a minimally invasive way. Though many factors have been shown to be involved in infertility, developing a profile of biomarkers that are consistent across infertility phenotypes remains elusive. As our understanding of the factors involved grows, it will become possible to implement our knowledge of these factors into the clinical decision-making process.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Larry I. Lipshultz, Alexander W. Pastuszak) for the focused issue “Contemporary Issues and Controversies in Men’s Health” published in Translational Andrology and Urology. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: The focused issue “Contemporary Issues and Controversies in Men’s Health” was commissioned by the editorial office without any funding or sponsorship. The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- World Health Organization, Department of Reproductive Health and Research. WHO laboratory manual for the examination and processing of human semen. Fifth edition. 2010.
- Niederberger C. Male Infertility. In: Campbell-Walsh Urology. Elsevier, Inc., 2016:556-79.e10.
- Smith RP, Coward RM, Lipshultz LL. The Office Visit. In: Current Management of Male Infertility. Elsevier, Inc., 2014:19-37.
- Guzick DS, Overstreet JW, Factor-Litvak P, et al. Sperm morphology, motility, and concentration in fertile and infertile men. N Engl J Med 2001;345:1388-93. [Crossref] [PubMed]
- Aston KI, Uren PJ, Jenkins TG, et al. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril 2015;104:1388-97.e1-5.
- Aston KI, Conrad DF. A review of genome-wide approaches to study the genetic basis for spermatogenic defects. Methods Mol Biol 2013;927:397-410. [Crossref] [PubMed]
- Hotaling JM. Genetics of Male Infertility. In: Current Management of Male Infertility. Elsevier, Inc., 2014:1-17.
- Hotaling J, Carrell DT. Clinical genetic testing for male factor infertility: current applications and future directions. Andrology 2014;2:339-50. [Crossref] [PubMed]
- Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med 2008;14:1197-213. [Crossref] [PubMed]
- MeSH Browser. Available online: (accessed 24 January 2019).https://meshb.nlm.nih.gov/record/ui?ui=D015415
- Magnus O, Tollefsrud A, Abyholm T, et al. Effects of varying the abstinence period in the same individuals on sperm quality. Arch Androl 1991;26:199-203. [Crossref] [PubMed]
- Sergerie M, Mieusset R, Croute F, et al. High risk of temporary alteration of semen parameters after recent acute febrile illness. Fertil Steril 2007;88:970.e1-7. [Crossref] [PubMed]
- Bhattacharya SM, Ghosh M, Nandi N. Diabetes mellitus and abnormalities in semen analysis. J Obstet Gynaecol Res 2014;40:167-71. [Crossref] [PubMed]
- La J, Roberts NH, Yafi FA. Diet and Men’s Sexual Health. Sex Med Rev 2018;6:54-68. [Crossref] [PubMed]
- Jóźków P, Mędraś M, Lwow F, et al. Associations between physical activity and semen quality in young healthy men. Fertil Steril 2017;107:373-8.e2. [Crossref] [PubMed]
- Gabrielsen JS, Tanrikut C. Chronic exposures and male fertility: the impacts of environment, diet, and drug use on spermatogenesis. Andrology 2016;4:648-61. [Crossref] [PubMed]
- Wijesekara GUS, Fernando DMS, Wijerathna S, et al. Environmental and occupational exposures as a cause of male infertility. Ceylon Med J 2015;60:52-6. [Crossref] [PubMed]
- Cui D, Han G, Shang Y, et al. Antisperm antibodies in infertile men and their effect on semen parameters: a systematic review and meta-analysis. Clin Chim Acta 2015;444:29-36. [Crossref] [PubMed]
- Oehninger S, Franken DR, Ombelet W. Sperm functional tests. Fertil Steril 2014;102:1528-33. [Crossref] [PubMed]
- Rex AS, Aagaard J, Fedder J. DNA fragmentation in spermatozoa: a historical review. Andrology 2017;5:622-30. [Crossref] [PubMed]
- Simon L, Emery BR, Carrell DT. Review: Diagnosis and impact of sperm DNA alterations in assisted reproduction. Best Pract Res Clin Obstet Gynaecol 2017;44:38-56. [Crossref] [PubMed]
- Bachir BG, Jarvi K. Infectious, Inflammatory, and Immunologic Conditions Resulting in Male Infertility. In: Current Management of Male Infertility. Elsevier, Inc., 2014:67-81.
- Francavilla F, Sciarretta F, Sorgentone S, et al. Intrauterine insemination with or without mild ovarian stimulation in couples with male subfertility due to oligo/astheno- and/or teratozoospermia or antisperm antibodies: a prospective cross-over trial. Fertil Steril 2009;92:1009-11. [Crossref] [PubMed]
- Leushuis E, van der Steeg JW, Steures P, et al. Immunoglobulin G antisperm antibodies and prediction of spontaneous pregnancy. Fertil Steril 2009;92:1659-65. [Crossref] [PubMed]
- Zini A, Fahmy N, Belzile E, et al. Antisperm antibodies are not associated with pregnancy rates after IVF and ICSI: systematic review and meta-analysis. Hum Reprod 2011;26:1288-95. [Crossref] [PubMed]
- Bungum M, Humaidan P, Axmon A, et al. Sperm DNA integrity assessment in prediction of assisted reproduction technology outcome. Hum Reprod 2007;22:174-9. [Crossref] [PubMed]
- Simon L, Zini A, Dyachenko A, et al. A systematic review and meta-analysis to determine the effect of sperm DNA damage on in vitro fertilization and intracytoplasmic sperm injection outcome. Asian J Androl 2017;19:80-90. [PubMed]
- O’Neill CL, Parrella A, Keating D, et al. A treatment algorithm for couples with unexplained infertility based on sperm chromatin assessment. J Assist Reprod Genet 2018;35:1911-7. [Crossref] [PubMed]
- Agarwal A, Majzoub A, Esteves SC, et al. Clinical utility of sperm DNA fragmentation testing: practice recommendations based on clinical scenarios. Transl Androl Urol 2016;5:935-50. [Crossref] [PubMed]
- Esteves SC, Roque M, Bradley CK, et al. Reproductive outcomes of testicular versus ejaculated sperm for intracytoplasmic sperm injection among men with high levels of DNA fragmentation in semen: systematic review and meta-analysis. Fertil Steril 2017;108:456-67.e1. [Crossref] [PubMed]
- Awaga HA, Bosdou JK, Goulis DG, et al. Testicular versus ejaculated spermatozoa for ICSI in patients without azoospermia: A systematic review. Reprod Biomed Online 2018;37:573-80. [Crossref] [PubMed]
- MeSH Browser. Available online: (accessed 28 January 2019).https://meshb.nlm.nih.gov/record/ui?ui=D023281
- Foote S, Vollrath D, Hilton A, et al. The human Y chromosome: overlapping DNA clones spanning the euchromatic region. Science 1992;258:60-6. [Crossref] [PubMed]
- Reijo R, Alagappan RK, Patrizio P, et al. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet 1996;347:1290-3. [Crossref] [PubMed]
- Reijo R, Lee TY, Salo P, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet 1995;10:383-93. [Crossref] [PubMed]
- Hopps CV, Mielnik A, Goldstein M, et al. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod 2003;18:1660-5. [Crossref] [PubMed]
- Jarow JP, Sigman M, Kolettis PN, et al. The Optimal Evaluation of the Infertile Male. AUA Best Practice Statement. 2010; discussion 25-6.
- Van Assche E, Bonduelle M, Tournaye H, et al. Cytogenetics of infertile men. Hum Reprod 1996;11 Suppl 4:1-24. [Crossref] [PubMed]
- Mayeur A, Ahdad N, Hesters L, et al. Chromosomal translocations and semen quality: A study on 144 male translocation carriers. Reprod Biomed Online 2019;38:46-55. [Crossref] [PubMed]
- Gao M, Pang H, Zhao YH, et al. Karyotype analysis in large sample cases from Shenyang Women’s and Children’s hospital: a study of 16,294 male infertility patients. Andrologia 2017. [Crossref] [PubMed]
- Hwang K, Weedin JW, Lamb DJ. The use of fluorescent in situ hybridization in male infertility. Ther Adv Urol 2010;2:157-69. [Crossref] [PubMed]
- Krausz C, Riera-Escamilla A. Genetics of male infertility. Nat Rev Urol 2018;15:369-84. [Crossref] [PubMed]
- Okutman O, Rhouma MB, Benkhalifa M, et al. Genetic evaluation of patients with non-syndromic male infertility. J Assist Reprod Genet 2018;35:1939-51. [Crossref] [PubMed]
- Cannarella R, Condorelli RA, Duca Y, et al. New insights into the genetics of spermatogenic failure: a review of the literature. Hum Genet 2019;138:125-40. [Crossref] [PubMed]
- Patel B, Parets S, Akana M, et al. Comprehensive genetic testing for female and male infertility using next-generation sequencing. J Assist Reprod Genet 2018;35:1489-96. [Crossref] [PubMed]
- MeSH Browser. Available online: (accessed 4 February 2019).https://meshb.nlm.nih.gov/record/ui?name=EPIGENETIC%20PROCESSES
- Bui AD, Sharma R, Henkel R, et al. Reactive oxygen species impact on sperm DNA and its role in male infertility. Andrologia 2018;50:e13012. [Crossref] [PubMed]
- Jenkins TG, Aston KI, James ER, et al. Sperm epigenetics in the study of male fertility, offspring health, and potential clinical applications. Syst Biol Reprod Med 2017;63:69-76. [Crossref] [PubMed]
- Denomme MM, McCallie BR, Parks JC, et al. Inheritance of epigenetic dysregulation from male factor infertility has a direct impact on reproductive potential. Fertil Steril 2018;110:419-28.e1. [Crossref] [PubMed]
- Aoki VW, Emery BR, Liu L, et al. Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl 2006;27:890-8. [Crossref] [PubMed]
- Santi D, De Vincentis S, Magnani E, et al. Impairment of sperm DNA methylation in male infertility: a meta-analytic study. Andrology 2017;5:695-703. [Crossref] [PubMed]
- Jenkins TG, Aston KI, Meyer TD, et al. Decreased fecundity and sperm DNA methylation patterns. Fertil Steril 2016;105:51-57.e1-3.
- García-Herrero S, Meseguer M, Martínez-Conejero JA, et al. The transcriptome of spermatozoa used in homologous intrauterine insemination varies considerably between samples that achieve pregnancy and those that do not. Fertil Steril 2010;94:1360-73. [Crossref] [PubMed]
- Levine H, Jørgensen N, Martino-Andrade A, et al. Temporal trends in sperm count:a systematic review and meta-regression analysis. Hum Reprod Update 2017;23:646-59. [Crossref] [PubMed]
- Menezo Y, Dale B, Elder K. The negative impact of the environment on methylation/epigenetic marking in gametes and embryos: A plea for action to protect the fertility of future generations. Mol Reprod Dev 2019;86:1273-82. [Crossref] [PubMed]
- Manikkam M, Tracey R, Guerrero-Bosagna C, et al. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One 2013;8:e55387. [Crossref] [PubMed]
- Katib A. Mechanisms linking obesity to male infertility. Cent European J Urol 2015;68:79-85. [Crossref] [PubMed]
- Craig JR, Jenkins TG, Carrell DT, et al. Obesity, male infertility, and the sperm epigenome. Fertil Steril 2017;107:848-59. [Crossref] [PubMed]
- Gunes S, Arslan MA, Hekim GNT, et al. The role of epigenetics in idiopathic male infertility. J Assist Reprod Genet 2016;33:553-69. [Crossref] [PubMed]
- Khawar MB, Mehmood R, Roohi N. MicroRNAs: Recent insights towards their role in male infertility and reproductive cancers. Bosn J Basic Med Sci 2019;19:31-42. [Crossref] [PubMed]
- Trigg NA, Eamens AL, Nixon B. The contribution of epididymosomes to the sperm small RNA profile. Reproduction 2019. [Epub ahead of print]. [Crossref] [PubMed]
- MeSH Browser. Available online: (accessed 4 February 2019).https://meshb.nlm.nih.gov/record/ui?ui=D040901
- Jodar M, Soler-Ventura A, Oliva R, et al. Semen proteomics and male infertility. J Proteomics 2017;162:125-34. [Crossref] [PubMed]
- Amaral A, Castillo J, Ramalho-Santos J, et al. The combined human sperm proteome: cellular pathways and implications for basic and clinical science. Hum Reprod Update 2014;20:40-62. [Crossref] [PubMed]
- Agarwal A, Bertolla RP, Samanta L. Sperm proteomics: potential impact on male infertility treatment. Expert Rev Proteomics 2016;13:285-96. [Crossref] [PubMed]
- Martínez-Heredia J, de Mateo S, Vidal-Taboada JM, et al. Identification of proteomic differences in asthenozoospermic sperm samples. Hum Reprod 2008;23:783-91. [Crossref] [PubMed]
- Amaral A, Ramalho-Santos J, St John JC. The expression of polymerase gamma and mitochondrial transcription factor A and the regulation of mitochondrial DNA content in mature human sperm. Hum Reprod 2007;22:1585-96. [Crossref] [PubMed]
- Liao TT, Xiang Z, Zhu WB, et al. Proteome analysis of round-headed and normal spermatozoa by 2-D fluorescence difference gel electrophoresis and mass spectrometry. Asian J Androl 2009;11:683-93. [Crossref] [PubMed]
- Xu W, Hu H, Wang Z, et al. Proteomic characteristics of spermatozoa in normozoospermic patients with infertility. J Proteomics 2012;75:5426-36. [Crossref] [PubMed]
- Behrouzi B, Kenigsberg S, Alladin N, et al. Evaluation of potential protein biomarkers in patients with high sperm DNA damage. Syst Biol Reprod Med 2013;59:153-63. [Crossref] [PubMed]
- Agarwal A, Sharma R, Durairajanayagam D, et al. Major protein alterations in spermatozoa from infertile men with unilateral varicocele. Reprod Biol Endocrinol 2015;13:8. [Crossref] [PubMed]
- Sharma R, Agarwal A, Mohanty G, et al. Proteomic analysis of human spermatozoa proteins with oxidative stress. Reprod Biol Endocrinol 2013;11:48. [Crossref] [PubMed]
- Parte PP, Rao P, Redij S, et al. Sperm phosphoproteome profiling by ultra performance liquid chromatography followed by data independent analysis (LC-MS(E)) reveals altered proteomic signatures in asthenozoospermia. J Proteomics 2012;75:5861-71. [Crossref] [PubMed]
- Blickenstorfer K, Voelkle M, Xie M, et al. Are WHO Recommendations to Perform 2 Consecutive Semen Analyses for Reliable Diagnosis of Male Infertility Still Valid? J Urol 2019;201:783-91. [Crossref] [PubMed]
- Chen Z, Toth T, Godfrey-Bailey L, et al. Seasonal variation and age-related changes in human semen parameters. J Androl 2003;24:226-31. [Crossref] [PubMed]
- Duncan MW, Thompson HS. Proteomics of semen and its constituents. Proteomics Clin Appl 2007;1:861-75. [Crossref] [PubMed]
- Buñay J, Larriba E, Patiño-Garcia D, et al. Combined proteomic and miRNome analyses of mouse testis exposed to an endocrine disruptors chemicals mixture reveals altered toxicological pathways involved in male infertility. Mol Hum Reprod 2019;25:156-69. [Crossref] [PubMed]
- Barrachina F, Jodar M, Delgado-Dueñas D, et al. Novel and conventional approaches for the analysis of quantitative proteomic data are complementary towards the identification of seminal plasma alterations in infertile patients. Mol Cell Proteomics 2019;18:S77-90. [Crossref] [PubMed]
- Dias TR, Samanta L, Agarwal A, et al. Proteomic Signatures Reveal Differences in Stress Response, Antioxidant Defense and Proteasomal Activity in Fertile Men with High Seminal ROS Levels. Int J Mol Sci 2019. [Crossref] [PubMed]
- Liu X, Wang X, Liu F. Decreased expression of heat shock protein A4L in spermatozoa is positively related to poor human sperm quality. Mol Reprod Dev 2019;86:379-86. [Crossref] [PubMed]
- MeSH Browser. Available online: (accessed 4 February 2019).https://meshb.nlm.nih.gov/record/ui?ui=D055432
- ter Kuile BH, Westerhoff HV. Transcriptome meets metabolome: hierarchical and metabolic regulation of the glycolytic pathway. FEBS Lett 2001;500:169-71. [Crossref] [PubMed]
- Kovac JR, Pastuszak AW, Lamb DJ. The use of genomics, proteomics, and metabolomics in identifying biomarkers of male infertility. Fertil Steril 2013;99:998-1007. [Crossref] [PubMed]
- Zhang Z, Zhang Y, Liu C, et al. Serum Metabolomic Profiling Identifies Characterization of Non-Obstructive Azoospermic Men. Int J Mol Sci 2017. [Crossref] [PubMed]
- Gilany K, Jafarzadeh N, Mani-Varnosfaderani A, et al. Metabolic Fingerprinting of Seminal Plasma from Non-obstructive Azoospermia Patients: Positive Versus Negative Sperm Retrieval. J Reprod Infertil 2018;19:109-14. [PubMed]
- Gilany K, Mani-Varnosfaderani A, Minai-Tehrani A, et al. Untargeted metabolomic profiling of seminal plasma in nonobstructive azoospermia men: A noninvasive detection of spermatogenesis. Biomed Chromatogr 2017. [Crossref] [PubMed]
- Zhao K, Zhang J, Xu Z, et al. Metabolomic Profiling of Human Spermatozoa in Idiopathic Asthenozoospermia Patients Using Gas Chromatography-Mass Spectrometry. Biomed Res Int 2018;2018:8327506.
- Qiao S, Wu W, Chen M, et al. Seminal plasma metabolomics approach for the diagnosis of unexplained male infertility. PLoS One 2017;12:e0181115. [Crossref] [PubMed]
- Guo D, Wu W, Tang Q, et al. The impact of BMI on sperm parameters and the metabolite changes of seminal plasma concomitantly. Oncotarget 2017;8:48619-34. [PubMed]
- Lefèvre PLC, Palin MF, Murphy BD. Polyamines on the reproductive landscape. Endocr Rev 2011;32:694-712. [Crossref] [PubMed]


