
Development and validation of a novel 5 cuproptosis-related long noncoding RNA signature to predict diagnosis, prognosis, and drug therapy in clear cell renal cell carcinoma
Highlight box
Key findings
• We systematically explored the role of cuproptosis in clear cell renal cell carcinoma (ccRCC). The cuproptosis gene is extensively deregulated in ccRCC. Furthermore, we constructed a signature consisting of 5 cuproptosis-related long noncoding RNAs (CRL) which could more accurately predict the diagnosis, prognosis, and clinical treatment of patients.
What is known and what is new?
• ccRCC is the main type of renal cell carcinoma. Cell death plays an important role in the development of tumors. Cuproptosis has attracted much attention as a novel cell death mode;
• In this study, a signature was constructed, and the values of the signature were verified, with the expression of key CRLs was verified by in vitro experiments.
What is the implication, and what should change now?
• The results showed that this signature could effectively predict the clinical characteristics of patients and help promote accurate diagnosis and treatment. Next, we hope to conduct more validation of signature effectiveness using clinical samples collected by ourselves to improve our study.
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common type of renal cell carcinoma, which is considered to originate from renal tubular epithelial cells. It is the third largest tumor disease in the urinary system, and the incidence is increasing each year (1,2). At present, there are a series of treatments for ccRCC, including surgical resection, targeted drug therapy, and new immunotherapy (3). For surgical treatment, 30% of patients with localized ccRCC (i.e., stage I–III) develop recurrence and metastasis after surgical tumor resection (4). For drug treatment, different patients have different responses to treatment. Therefore, it is essential to explore the potential mechanism of ccRCC, identify new biomarkers for different populations, and construct more effective prognostic targets for diagnosis and treatment.
Copper is one of the key trace elements in human body, and it is also a cofactor of many key enzymes in cells. Rae et al. found that there are specific mechanisms to regulate the homeostasis of copper ions in cells (5). Kim et al. reviewed recent developments and found that copper ions were toxic to the body when they exceeded a certain threshold (6). Ge et al.’s literature review found that patients with cancer have higher levels of copper in serum and tissue samples, and intracellular copper levels can affect the progress of cancer, suggesting that copper ions play an important role in the occurrence and development of cancer (7). Recently, Tsvetkov et al. proposed a new type of cell death called cuproptosis (8). This cell death mode does not depend on the previously studied signaling pathways such as apoptosis (9), cell necrosis (10), cell pyroptosis (11), or ferroptosis (12). The mechanism involves copper ions directly binding to the fatty acylation components of the tricarboxylic acid cycle (TCA), resulting in toxic protein stress and ultimately cell death (13). This is consistent with the previous finding of Wettersten et al. who reported that there is a reprogramming of related metabolic pathways in renal clear cell carcinoma (14).
Long noncoding RNA (lncRNA) is a noncoding RNA of more than 200 nucleotides in length (15). It affects gene transcription and translation through a variety of biological processes. A growing body of evidence indicates that lncRNA plays an important role in the development and prognosis of tumors (16). Xue et al. found that the mitotically-associated lncRNA MANCR could play an important influence on the progression of ccRCC by constructing a prognostic lncRNA model (17). In addition, Zhang et al. found that the cuproptosis-associated lncRNA signature could be used to predict the immune infiltration and prognosis of bladder cancer patients by systematically analyzing the expression characteristics of cuproptosis-associated lncRNA (18). However, the mechanism of cuproptosis gene and its related lncRNA in ccRCC remains unclear.
In the present study, we constructed an assessment signature containing 5 cuproptosis-related lncRNAs (CRLs) and evaluated its value for the diagnosis, prognosis prediction, and clinical treatment of ccRCC. The results showed that the signature has great application potential and may provide guidance for clinical precision treatment. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-23-65/rc).
Methods
Expression profile and clinical data acquisition
The transcripts per kilobase per million (TPM) format gene expression matrix, nucleotide mutation, copy number variation (CNV) data, and clinical information data of ccRCC were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The gene matrix included 537 ccRCC tissues and 72 adjacent tissues. Clinical information included age, gender, clinical stage, nuclear grade, T stage, M stage, N stage, survival status, and survival time of 539 patients. R software (The R Foundation of Statistical Computing) was used to annotate the RNA sequencing (RNA-seq) data. For different expression values of the same gene, we selected the average value for analysis and then screened the expression matrix of messenger RNA (mRNA) and lncRNA. The gene expression matrix data were normalized by log transformation. The “limma” package (19) in R software was used to correct the gene expression matrix data we download.
Twenty pairs of tissue samples were collected from ccRCC patients who underwent partial/radical nephrectomy in the Department of Urology at the First Hospital of Shanxi Medical University from June 2021 to December 2021. These tissue samples were taken from postoperative specimens of these patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Hospital of Shanxi Medical University (No. 2021K034), and all patients provided written informed consent. The inclusion criteria for patients were the following: patients did not receive preoperative radiotherapy, chemotherapy, molecular targeted therapy, or other antitumor treatment; the diagnosis of ccRCC was confirmed by histopathology; and patient clinical information (e.g., age, sex, T stage, nuclear grade) was complete.
Analysis of cuproptosis-related gene expression characteristics
The expression matrix of 10 cuproptosis key genes was extracted from all RNA-seq expression matrix data. In order to study the function of cuproptosis key genes in ccRCC, the “limma” R package was used to identify the differential expression levels of cuproptosis key genes between ccRCC tissues and normal kidney tissues. P<0.05 was considered statistically significant. In R, the software packages “pheatmap” and “boxplot” packages were used to visualize the expression level and differences in cuproptosis gene, the “corrplot” package was used to conduct Spearman correlation analysis to determine the potential correlation of 10 cuproptosis-related genes, the “RCircos” and “maftools” packages were used to visualize the mutations in key genes of cuproptosis, and the “survival” package was used to detect the prognostic value of key differentially expressed cuproptosis genes.
Consensus clustering of CRLs
The CRLs related to prognosis were screened by Cox regression analysis for further analysis. The prognostic differentially expressed CRLs were screened by Wilcoxon signed rank test and Pearson correlation analysis. The threshold was set as correlation coefficient |R| >0.4 and P<0.001. In order to verify whether patients with ccRCC can be grouped by key CRLs, we performed consensus clustering by using R “ConsensusClusterPlus” package. The cell type identification by estimating relative subsets of RNA transcripts (CIBERSORT) and estimation of stromal and immune cells in malignant tumor tissues using expression data (ESTIMATE) algorithms were used to evaluate the immune cell infiltration and tumor microenvironment between different groups (20,21).
Construction of the CRL signature
To establish an effective prognostic model, 530 patients with ccRCC with survival data were randomly divided into a training cohort and testing cohort at a 1:1 ratio. Finally, 266 people were included in the training cohort, and 264 people were included in the testing cohort. The clinical features between groups can be found in Table 1. The lncRNA scoring formula related to cuproptosis was constructed through the training cohort, and the prognosis prediction ability of the scoring formula was evaluated through the testing cohort and TCGA total cohort. A lncRNA scoring formula was constructed by least absolute shrinkage and selection operator (LASSO) regression analysis. The formula was as follows: risk score = ∑Coef lncRNA × Exp lncRNA, where Coef is the regression coefficient.
Table 1
| Characteristics | Training cohort (N=266) | Testing cohort (N=264) | TCGA cohort (N=530) |
|---|---|---|---|
| Age (years), n (%) | |||
| ≤65 | 183 (17.3) | 165 (15.6) | 348 (32.8) |
| >65 | 83 (7.8) | 99 (9.3) | 182 (17.2) |
| Gender, n (%) | |||
| Female | 95 (9.0) | 92 (8.7) | 187 (17.6) |
| Male | 171 (16.1) | 172 (16.2) | 343 (32.4) |
| Grade, n (%) | |||
| G1–2 | 116 (10.9) | 126 (11.9) | 242 (22.8) |
| G3–4 | 145 (13.7) | 135 (12.7) | 280 (26.4) |
| Unknown | 5 (0.5) | 3 (0.3) | 8 (0.8) |
| Stage, n (%) | |||
| Stage I–II | 161 (15.2) | 162 (15.3) | 323 (30.5) |
| Stage III–IV | 102 (9.6) | 102 (9.6) | 204 (19.2) |
| Unknown | 3 (0.3) | 0 (0.0) | 3 (0.3) |
| T stage, n (%) | |||
| T1–2 | 171 (16.1) | 170 (16.0) | 341 (32.2) |
| T3–4 | 95 (9.0) | 94 (8.9) | 189 (17.8) |
| M stage, n (%) | |||
| M0 | 210 (19.8) | 210 (19.8) | 420 (39.6) |
| M1 | 40 (3.8) | 38 (3.6) | 78 (7.4) |
| Unknown | 16 (1.5) | 16 (1.5) | 32 (3.0) |
| N stage, n (%) | |||
| N0 | 113 (10.7) | 126 (11.9) | 239 (22.5) |
| N1 | 7 (0.7) | 9 (0.8) | 16 (1.5) |
| Unknown | 146 (13.8) | 129 (12.2) | 275 (25.9) |
ccRCC, clear cell renal cell carcinoma; TCGA, The Cancer Genome Atlas.
The prognostic value of the CRL signature
Univariate Cox and multivariate Cox regression analyses were used to evaluate whether the scoring formula could be used as an independent prognostic factor. The patients were divided into a low-risk group and a high-risk group according to the median risk score, and the overall survival (OS) between the 2 groups was compared by log-rank test. T-distributed stochastic neighbor embedding (t-SNE) was used to visualize the distribution characteristics of different risk groups. The stability of the model was evaluated by the testing cohort and TCGA cohort. The “ROC” R package was used to draw the receiver operating characteristic (ROC) curve, and the area under the curve (AUC) showed the specificity and sensitivity of the model.
Construction of the prognostic nomogram
By combining the risk score and prognostic-related clinical variables (age, grade, and stage), we used R software “rms” package to construct a prognostic map that could predict the 1-, 3-, and 5-year survival rates of patients with ccRCC. We evaluated the reliability and accuracy of the plot by calibration curve, ROC, curve and, decision curve analysis (DCA) analysis.
The tumor microenvironment prediction value of the CRL signature
According to the median risk score, patients with ccRCC were divided into a low-risk group and high-risk group. Gene set enrichment analysis (GSEA) software (version 4.1.0; http://www.broad.mit.edu/gsea/) was used to analyze the differential pathways between the high- and low-risk groups. Thresholds were set as adjusted P<0.05 and false discovery rate (FDR) <0.25. The activity of 13 immune-related pathways was calculated by single-sample GSEA (ssGSEA) using the “GSVA” package in R, while the “limma” package was used to analyze the expression differences of immune checkpoint genes between the high- and low-risk groups. The CIBERSORT algorithm was used to calculate the level of immune cell infiltration, and Pearson correlation analysis was used to detect the correlation between the risk score and immune cell infiltration level.
The clinical therapy prediction value of the CRL signature
In order to evaluate the role of the predictive risk score in predicting the clinical treatment of patients with ccRCC, we analyzed the expression levels of key genes that may affect the efficacy of immunosuppressant programmed cell death protein 1 (PD-1) in the high- and low-risk groups, and used the R package “pRRophetic” to predict the half maximum inhibitory concentration (IC50) of the common targeted drugs in clinical treatment (22). The Wilcoxon signed-rank test was used to compare the IC50 values between the low-risk group and the high-risk group.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The clinical tissues were immediately transferred to RNA protection solution after resection. Total RNA was extracted from tissues and cell lines with the TRIzol method. A Droplight304 (Azanno Biotech) spectrometer was used to determine RNA concentration and purity. The RNA template was reversely transcribed using a PCR kit (AUQ01; TransGen Biotech) to detect the expression levels of 5 lncRNAs (AC015912.3, AC026401.3, AC103706.1, AC134312.5, EMX2OS) and the internal reference gene GAPDH. Quantitative PCR was performed using a real-time fluorescent quantitative PCR instrument (7500 Fast Dx Real-Time PCR, Applied Biosystems, Thermo Fisher Scientific). Primers designed with National Center for Biotechnology Information (NCBI) Primer Basic Local Alignment Search Tool (BLAST) are shown in Table 2.
Table 2
| Gene | Forward primer | Reverse primer |
|---|---|---|
| AC015912.3 | CTGAAGCCGTCAGGAAGTCA | GGTCGCGAGGATGCTCTAAA |
| AC026401.3 | GAATTACGCTGCGATGGTGG | AAGCCTCTTTGACCAGAAGCC |
| AC103706.1 | TGCAGTTATTCAGCCCTCCC | TTAGCATCAAAGCGCTGCAC |
| AC134312.5 | TGACCTGGGAAAAGCCCATC | AGAACAGATGCAGCACGGAA |
| EMX2OS | ATCCCTCCTCAGAACCCCTC | AAACATGCAAAGACCGTGCC |
| GAPDH | GTCCACCACCCTGTTGCTGTA | ACCCACTCCTCCACCTTTGA |
Statistical analysis
All statistical analyses were performed with R software (version 4.1.3). The expression differences of key cuproptosis genes and lncRNAs in cancer tissues and paired adjacent normal tissues were analyzed by t-test. Univariate Cox regression analysis was used to identify the prognostic value of key genes and lncRNAs in cuproptosis. LASSO regression analysis was used to screen key lncRNAs in cuproptosis to construct a predictive scoring formula. Kaplan-Meier method and log-rank test were used to analyze the survival rate of patients in different groups. R software was used to perform heatmap, GSEA, survival curve, ROC curve, plot, calibration curve, and DCA curve analyses. P<0.05 was considered statistically significant.
Results
Expression characteristics of cuproptosis-related genes in ccRCC
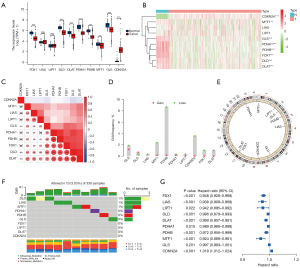
The analysis flowchart of this study is shown in Figure 1. First, we analyzed the expression of 10 cuproptosis-related genes in TCGA cohort. The results showed that cuproptosis-related genes were generally dysregulated in ccRCC, among which the CDKN2A gene was upregulated in tumor tissues, and FDX1, DLD, DLAT, PDHA1, PDHB, MTF1, and GLS were downregulated in tumor tissues (Figure 2A). We also drew a heatmap of gene expression (Figure 2B) which showed the same pattern. Subsequently, we performed correlation analysis on the expression trend of cuproptosis-related genes, and the results showed that most genes showed a positive correlation, among which DLAT had the strongest correlation with DLD (correlation =0.85), and CDKN2A had a negative correlation with the other genes (Figure 2C). Then we sorted out the CNV information of 380 cases of patients with ccRCC in TCGA database. The results showed that there were varying degrees of CNV in cuproptosis-related genes, and most of them had copy number deletion. Among them, the PDHB gene had the highest frequency of copy number deletion, which could reach 8%, but the frequency of copy number amplification was small. Among them, the GLS gene had the highest frequency of copy number amplification, but at less than 2% (Figure 2D). The location of the genes in the chromosome is shown in Figure 2E. In addition, we also analyzed the single-nucleotide polymorphism (SNP) of 330 patients with ccRCC in TCGA database. The results showed that 10 patients had mutations in the key genes of cuproptosis, most of which were deletion mutations, and the frequency of the DLD mutation was the highest (Figure 2F). In addition, combined with the clinical data of patients, we studied the prognostic value of 8 genes related to cuproptosis in patients with ccRCC. The results showed that all genes, except the GLS gene, showed good prognostic predictive ability (Figure 2G).
Key CRL screening
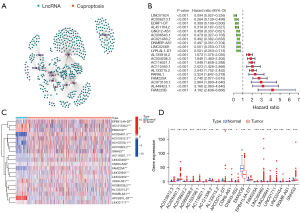
Pearson correlation analysis was used to screen 465 differentially expressed CRLs (Figure 3A) in TCGA data set. Subsequently, we analyzed the prognosis of the 465 CRLs, and 153 CRLs (Table S1) were screened and found to be significantly correlated with the survival prognosis. We constructed a forest map of the top 10 positive and top 10 negative correlation genes (Figure 3B). In addition, we also identified the top 20 lncRNAs closely related to survival and prognosis using a heatmap and differential expression map, with the results indicating significant differences (Figure 3C,3D).
Consensus clustering analysis based on the key CRLs
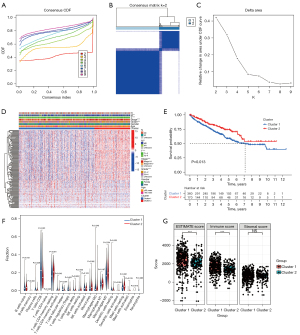
According to the expression similarity of 153 selected key CRLs, we conducted consensus clustering analysis on 530 patients with ccRCC and found that the cumulative distribution function (CDF) value was small when k=2 (Figure 4A-4C). Subsequently, we performed clinical correlation analysis on the consistent clustering results. The results showed that different clustering groups were closely related to a variety of clinical traits of patients, including stage and grade of tumors. Moreover, the prognosis of patients with different clustering groups was also significantly different (P=0.013; Figure 4D,4E). In addition, we used the CIBERSORT algorithm to evaluate the immune cell infiltration level of consistent clustering results. We found that there were a variety of immune cell infiltration differences across the different clustering groups, among which CD8 T cells, resting CD4 memory T cells, and M2 macrophages had higher infiltration levels and significant differences across the different groups (Figure 4F). In addition, we used the ESTIMATE algorithm to analyze the tumor microenvironment of the different clustering groups. The results showed that there were differences in tumor microenvironment scores and immune scores between the different clustering groups. Interestingly, we found that there was no significant difference in matrix scores between the 2 clustering groups (Figure 4G). These results show that the grouping identified by key CRLs may provide new predictive indicators for the clinical and prognostic prediction of patients, which warrants further study.
Construction of a CRL signature and prognostic evaluation
A total of 530 patients with ccRCC were randomly divided into a training cohort (n=266) and a testing cohort (n=264). We then used the training cohort to perform LASSO regression analysis on 153 CRLs and constructed a CRL signature containing 5 CRLs (Table S2). The scoring method was as follows: risk score = AC015912.3 × 0.011 + AC026401.3 × 0.015 + AC103706.1 × 0.068 + AC134312.5 × 0.103 – EMX2OS × 0.011. The risk score of each patient was calculated according to the 5-CRL signature. According to the median risk score, patients with ccRCC were divided into a low-risk group and a high-risk group. Kaplan-Meier survival analysis showed that patients with ccRCC in the low-risk group had better clinical prognosis than did those in the high-risk group. The training cohort (Figure 5A), testing cohort (Figure 5B), and total TCGA cohort (Figure S1A) showed significant differences in prognosis (training cohort P<0.001; testing cohort P=0.007; total TCGA cohort P<0.001). In addition, we used the ROC curve to predict the accuracy of the 5-CRL signature. The results showed that the signature could accurately predict the survival rate of patients in the training cohort, testing cohort, and the total TCGA cohort (Figure 5C,5D; Figure S1B). In addition, the risk assessment results show that the survival state of patients with a high score was poor (Figure 5E,5F; Figure S1C). The t-SNE analysis results showed that there were significant differences between the high- and low-risk patients as distinguished by risk score (Figure 5G,5H; Figure S1D). Combined with the clinical parameters of age, gender, tumor stage, and grade of patients with ccRCC in TCGA data cohort, we conducted univariate and multivariate Cox analysis, and the results showed that the 5-CRL signature related to cuproptosis could be used as an independent prognostic factor for patients (Figure 5I,5J). These conclusions were verified in the testing cohort and the total TCGA cohort (Figure 5K,5L; Figure S1E,S1F; Table S3).

Correlation between the 5-CRL signature and clinical characteristics
After studying the relationship between risk score and clinical parameters, we found that the risk score was closely related to the stage, grade, and TNM status of patients with ccRCC, and high risk was usually accompanied by higher stage, grade, and TNM status (Figure 6A). In addition, compared with those in the low-risk group, patients with ccRCC in the high-risk group usually had a higher proportion of AC015912.3, AC026401.3, AC103706.1, and AC134312.5 (Figure 6A). We then analyzed whether there were differences in risk scores among patients with different clinical traits. The results showed there were significant differences in risk scores among tumor patients with different genders, grading stages, staging grades, and TNM stages, but not for age (Figure 6B-6H; Figure S2). Among them, the risk scores of male patients were generally higher, and the risk scores of patients with higher grades and stages were higher. We also found that people with higher immune infiltration scores tended to have higher risk scores. These results showed that the risk score was closely related to the clinical characteristics of patients with ccRCC, suggesting that the signature may be a key indicator for clinical prediction of patients with ccRCC.
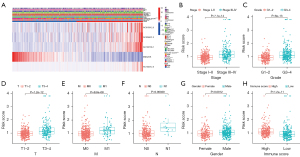
Stratified prognostic value of the 5 CRL signature
We assessed whether the prognostic value of the 5-CRL signature was still applicable in clinical parameter groups stratified by gender (female and male), age (≤65 and >65 years), stage (I–II and III–IV), grade (I–II and III–IV), T stage (T1–2 and T3–4), M stage (M0 and M1), and N stage (N0 and N1). The results showed that in most clinical parameter groups, there were significant differences in survival prognosis between the high- and low-risk patients. Patients in the high-risk group had worse survival prognosis, but there was no significant difference between low-grade, N1, and M1 between the groups (Figure 7; Figure S3).
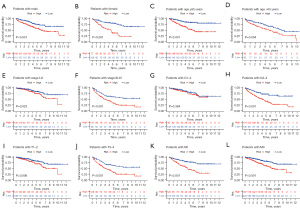
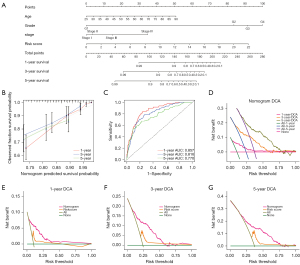
Construction and verification of the prognostic nomogram
By combining the clinical parameters closely related to the prognosis of patients, including gender, grade, stage, and the 5-CRL signature, we created a clinically applicable prediction tool to improve the accuracy of predicting the survival rate of patients with ccRCC. The nomogram was used to predict the 1-, 3-, and 5-year survival rates of patients with ccRCC (Figure 8A). Subsequently, we demonstrated the predictive ability of the prediction tool for 1-, 3-, and 5-year survival rates through calibration curves and ROC. The results showed that the nomogram had excellent prognostic accuracy for 1-, 3-, and 5-year survival rates (the AUCs were 0.857, 0.816, and 0.770, respectively; Figure 8B,8C). Furthermore, we applied DCA to evaluate the clinical applicability of the prediction tool more accurately. The results of DCA showed that the nomogram had better net benefit and a wider threshold probability range for predicting the 1-, 3-, and 5-year survival rates of patients. In addition, the nomogram provided better clinical benefit than did the 5-CRL signature (Figure 8D-8G).
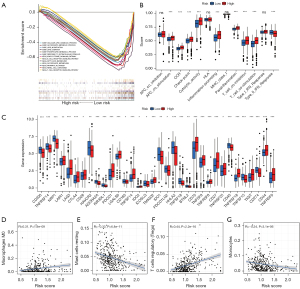
Evaluation of tumor immunity based on the 5 CRL signature
The patients with high and low scores were grouped, and the function analysis of the genes between groups was carried out with GSEA software. The function analysis results showed that the lipid metabolism pathway in the high-risk group was suppressed, suggesting that there might be metabolic abnormalities in the high-risk group. Meanwhile, the receptor signaling pathways of T cells and B cells were activated in the low-risk group, suggesting that there might be abnormal immune-related functions in the high-risk group (Figure 9A). Based on the differences in immune-related functions between the high- and low-risk groups, we further analyzed the differences in immune-related functions, immune cell infiltration, and the tumor microenvironment between patients with high- and low-risk scores. First, we analyzed the enrichment level and activity of immune cells, pathways, and functions in ccRCC. The results showed that there were significant differences in the expression of various immune indexes between the low-risk group and the high-risk groups (Figure 9B). Subsequently, we further analyzed the expression levels of immune checkpoints between the low-risk group and the high-risk group. By retrieving the expression of 34 key immune checkpoints between the high-risk group and the low-risk group, we found that there were differences in multiple immune checkpoint molecules between the groups (Figure 9C). In addition, we also studied the correlation between the risk score and immune cell infiltration. The results showed that memory B cells, M0 macrophages, plasma cells, activated CD4 memory T cells, CD8 T cells, regulatory T cells, and follicular helper T cells were positively correlated with the risk score. M1 macrophages, M2 macrophages, resting mast cells, monocytes, resting natural killer (NK) cells, resting CD4 memory T cells, eosinophils, activated dendritic cells, and resting dendritic cells were negatively correlated with the risk score (Figure 9D-9G, Figure S4).
Evaluation of the drug sensitivity of patients based on the 5-CRL signature
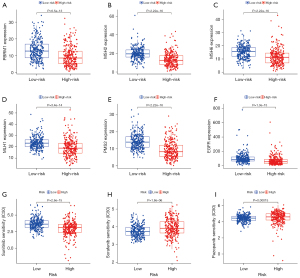
Based on the abnormality of multiple immune checkpoint molecules between the high- and low-risk patients, we analyzed the sensitivity of these patients to clinical treatment. First, we analyzed the expression of gene targets that may affect the therapeutic effect of the immune checkpoint inhibitor (ICI) PD-1 in high- and low-risk patients. The results showed that the expression level of PBRM1, a treatment-related sensitivity gene, was low in the high-risk group, suggesting that the high-risk group might receive a better therapeutic effect from PD-1 (Figure 10A). The expression levels of MSH2, MSH6, MLH1, and PMS2, a DNA mismatch repair-related gene, were significantly lower in the high-risk group, suggesting that patients could receive a better therapeutic benefit from PD-1 (Figure 10B-10E). The expression of EGFR in the treatment-related burst gene was lower in the high-risk group (Figure 10F), suggesting that PD-1 treatment was safer. We also analyzed the sensitivity of high-risk and low-risk patients to commonly used clinical targeted drugs. The results showed that the high-risk group was more sensitive to sunitinib (Figure 10G) while the low-risk group was more sensitive to sorafenib and pazopanib (Figure 10H,10I). These results show that the 5-CRL signature can be used as an independent prognostic indicator and may have important clinical application value.
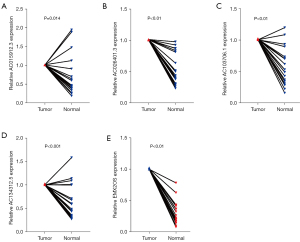
Tumor tissue validation
Twenty pairs of ccRCC and adjacent normal tissue samples were verified by qRT-PCR. The results showed that the relative expression of AC015912.3, AC026401.3, AC103706.1, and AC134312.5 in renal cell carcinoma was significantly higher than that in normal renal tissue, and the relative expression of EMX2OS in renal cell carcinoma was significantly lower than that in adjacent normal tissue (P<0.001; Figure 11). This suggests that AC015912.3, AC026401.3, AC103706.1, and AC134312.5 are cancer-promoting factors, while EMX2OS is a tumor-suppressor factor. These results are consistent with our previous analysis, but more samples are needed to verify these suppositions.
Discussion
ccRCC is one of the most common malignant tumors in the urinary system. In recent years, tremendous progress has been made in the treatment of ccRCC, and breakthroughs have been made in surgery and drug therapy (23). However, the early diagnosis, prognostic prediction, and clinical effect in ccRCC still require further study. Recently, cuproptosis has been discovered to be a novel type of cell death regulation, similar to ferroptosis. This pattern has been found to influence the progression of cancer disease. A review by Jiang et al. showed that copper ions can exert anti-tumor effects, suggesting that copper death may play a key role in the treatment of tumor diseases (24). Some studies have suggested that copper death-related genes play a key role in predicting clinical outcome in renal clear cell carcinoma (25,26). However, few studies have investigated the function of cuproptosis-related CRLs in ccRCC. In this paper, a 5-CRL signature was established based on these lncRNAs, and its prediction efficiency was evaluated.
In this study, we first examined the expression and mutation characteristics of 10 key cuproptosis genes in ccRCC in TCGA cohort. We found that cuproptosis genes were extensively dysregulated in ccRCC tumors, and most genes were significantly correlated with the prognosis of patients. Subsequently, we screened 153 characteristic prognostic lncRNAs related to cuproptosis genes by combining correlation analysis and Cox prognostic analysis. After the consensus clustering of patients with ccRCC using 153 key CRLs, we found that it was productive to use key CRLs to divide patients with ccRCC into 2 clusters. Subsequently, we used LASSO regression to screen the key CRLs and then identified and constructed the signature containing 5 lncRNAs. Patients with ccRCC were divided into a low-risk group and high-risk group with the 5-CRL signature. The results showed that the total survival time of patients in the high-risk group was shorter than that of patients in the low-risk group. The results of univariate and multivariate Cox analyses also suggested that the scoring formula could be used as an independent risk factor for predicting survival results. These results were verified in the testing cohort and TCGA total cohort, suggesting that the scoring formula could be used as a prognostic indicator. In addition, by analyzing the relationship between different clinical parameters and the 5-CRL signature, we found that there were significant differences in clinical parameters between the high- and low-risk groups, suggesting that the signature could be used as an indicator for clinical prediction. In addition, the 5-CRL signature has also been shown to be associated with OS in different clinical subgroups of ccRCC, including those of age, gender, grading, staging, T stage, N stage, and M stage. We thus established a nomogram including the risk score to calculate the 1-, 3-, and 5-year survival rates of patients with ccRCC, aiming to predict the prognosis of patients with ccRCC more accurately. The calibration curve and ROC suggested that the line chart had high accuracy, and the DCA decision curve suggested that it had higher sensitivity.
Cuproptosis-related lncRNAs have been found to be important in assessing immune function and tumor microenvironment in ccRCC patients. Chi et al. found that cuproptosis-related lncRNAs can predict the immune microenvironment of patients with Pancreatic adenocarcinoma (27). Therefore, we examined the power of the 5-CRLs signature to detect the immune characteristics of ccRCC patients. GSEA function enrichment analysis between the high- and low-risk groups showed that there were significant differences in in the levels of multiple metabolic pathways among different populations. Interestingly, there were also significant differences in immune function. The ssGSEA results suggest that there are differences in various immune characteristics among different risk populations, such as immune checkpoints. Therefore, we analyzed the levels of immune checkpoint expression across groups and found that there were significant differences in immune checkpoint expression. The correlation analysis between immune infiltration and risk score showed that macrophages M0 and Tregs cells were significantly positively correlated with risk score. This suggests that the high-risk group has more dormant macrophages and more immunosuppressive Tregs. These analyses suggest that patients in the high-risk group may have a higher probability of immune escape, thereby contributing to the development of neoplastic disease.
Cuproptosis-related lncRNAs have also been identified as important in guiding clinical treatment of tumor patients. Chen et al. found that cuproptosis-related lncRNAs can predict drug therapy response in hepatocellular carcinoma (28). Through this feature, we identified the differences in the sensitivity of patients in the high- and low-risk groups to common chemotherapy and immunotherapy drugs. A clinical trial study has reported that patients with the PBRM1 gene mutation are more sensitive to PD-1 inhibitors, resulting in significantly improved therapeutic effect (29). This study found that PBRM1 gene expression was lower in high-risk group, suggesting that the high-risk group may be more sensitive to PD-1 treatment. One study has shown that DNA mismatch repair genes play an important role in the ICI therapy of ccRCC (30). The results showed that there were significant differences in DNA mismatch repair genes between the different risk groups. In addition, a study has found that EGFR amplification can cause the outbreak of PD-1 treatment (31). In our study, the proportion of EGFR in the high-risk groups was significantly lower, indicating that PD-1 treatment in high-risk groups is safer. In addition, by studying the targeted chemotherapy drug sensitivity between high- and low-risk groups, we found that there were also significant differences. The high-risk population is sensitive to sunitinib, and the low-risk population is more sensitive to sorafenib and pazopanib, which provides the basis for the selection of clinical chemotherapy strategy. Subsequently, we verified the results through experiments, which showed that AC015912.3, AC026401.3, AC103706.1 and AC134312.5 were highly expressed in tumor tissues, while EMX2OS was poorly expressed in tumor tissues. Wang et al. found that lncRNA can enhance the resistance of hepatocellular carcinoma to targeted drugs through related pathways (32). Chen et al. confirmed that overexpression of EMX2OS can inhibit the growth of Wilms’ tumor in vitro (33). These findings suggest that the key lncRNAs we have identified may play an important role in the progression of a variety of tumors, but the regulatory mechanisms in ccRCC need further study.
Cuproptosis is a newly discovered type of cell death, the understanding of which can provide new insights for tumor research. However, our research has some limitations. Since we did not find a large-sample data sets with clinical data and gene expression, we only use TCGA database for model construction and validation. Therefore, the applicability of the 5-CRL signature needs to be tested using other public data sets or data sets collected by ourselves. In addition, the mechanism of key lncRNA contained in the 5-CRL signature in ccRCC needs to be verified by more in vivo and in vitro experiments in the future.
Conclusions
In this study, based on the expression characteristics of cuproptosis genes, we further identified CRLs, and constructed a 5-CRL signature. The results show that the signature can be used as an indicator to predict the diagnosis and prognosis of patients with ccRCC. It provides direction for the potential regulatory mechanisms of ccRCC and new horizons for clinical immunological drugs and targeted drug therapy strategies for patients with ccRCC.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 81970662), the Fundamental Research Program of Shanxi Province (No. 201901D211477), the Beijing Bethune Charitable Foundation (No. mnz1202029), and the Returned Overseas Students Scientific Research Funding Project of Shanxi Province (No. 2021-160).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-23-65/rc
Data Sharing Statement: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-65/dss
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-23-65/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-23-65/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the First Hospital of Shanxi Medical University (No. 2021K034) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Cao W, Chen HD, Yu YW, et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134:783-91. [Crossref] [PubMed]
- Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2022 Update. Eur Urol 2022;82:399-410. [Crossref] [PubMed]
- Li QK, Pavlovich CP, Zhang H, et al. Challenges and opportunities in the proteomic characterization of clear cell renal cell carcinoma (ccRCC): A critical step towards the personalized care of renal cancers. Semin Cancer Biol 2019;55:8-15. [Crossref] [PubMed]
- Rae TD, Schmidt PJ, Pufahl RA, et al. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 1999;284:805-8. [Crossref] [PubMed]
- Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol 2008;4:176-85. [Crossref] [PubMed]
- Ge EJ, Bush AI, Casini A, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer 2022;22:102-13. [Crossref] [PubMed]
- Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022;375:1254-61. [Crossref] [PubMed]
- Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol 2020;17:395-417. [Crossref] [PubMed]
- Weinlich R, Oberst A, Beere HM, et al. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol 2017;18:127-36. [Crossref] [PubMed]
- Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 2009;7:99-109. [Crossref] [PubMed]
- Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. [Crossref] [PubMed]
- Kahlson MA, Dixon SJ. Copper-induced cell death. Science 2022;375:1231-2. [Crossref] [PubMed]
- Wettersten HI, Aboud OA, Lara PN Jr, et al. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol 2017;13:410-9. [Crossref] [PubMed]
- Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol 2017;1008:1-46. [Crossref] [PubMed]
- Chen W, Yang J, Fang H, et al. Relevance Function of Linc-ROR in the Pathogenesis of Cancer. Front Cell Dev Biol 2020;8:696. [Crossref] [PubMed]
- Xue S, Guo H, Wei S, et al. Identification of a combined lncRNA prognostic signature and knockdown of lncRNA MANCR to inhibit progression of clear cell renal cell carcinoma by bioinformatics analysis. Transl Androl Urol 2022;11:1304-17. [Crossref] [PubMed]
- Zhang Y, Li X, Li X, et al. Comprehensive analysis of cuproptosis-related long noncoding RNA immune infiltration and prediction of prognosis in patients with bladder cancer. Front Genet 2022;13:990326. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9:e107468. [Crossref] [PubMed]
- Atkins MB, Tannir NM. Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat Rev 2018;70:127-37. [Crossref] [PubMed]
- Jiang Y, Huo Z, Qi X, et al. Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine (Lond) 2022;17:303-24. [Crossref] [PubMed]
- Bian Z, Fan R, Xie L. A Novel Cuproptosis-Related Prognostic Gene Signature and Validation of Differential Expression in Clear Cell Renal Cell Carcinoma. Genes (Basel) 2022;13:851. [Crossref] [PubMed]
- Wang T, Liu Y, Li Q, et al. Cuproptosis-related gene FDX1 expression correlates with the prognosis and tumor immune microenvironment in clear cell renal cell carcinoma. Front Immunol 2022;13:999823. [Crossref] [PubMed]
- Chi H, Peng G, Wang R, et al. Cuprotosis Programmed-Cell-Death-Related lncRNA Signature Predicts Prognosis and Immune Landscape in PAAD Patients. Cells 2022;11:3436. [Crossref] [PubMed]
- Chen S, Liu P, Zhao L, et al. A novel cuproptosis-related prognostic lncRNA signature for predicting immune and drug therapy response in hepatocellular carcinoma. Front Immunol 2022;13:954653. [Crossref] [PubMed]
- Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018;359:801-6. [Crossref] [PubMed]
- Møller P, Seppälä TT, Bernstein I, et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the Prospective Lynch Syndrome Database. Gut 2018;67:1306-16. [Crossref] [PubMed]
- Sugiyama E, Togashi Y, Takeuchi Y, et al. Blockade of EGFR improves responsiveness to PD-1 blockade in EGFR-mutated non-small cell lung cancer. Sci Immunol 2020;5:eaav3937. [Crossref] [PubMed]
- Wang Y, Tan K, Hu W, et al. LncRNA AC026401.3 interacts with OCT1 to intensify sorafenib and lenvatinib resistance by activating E2F2 signaling in hepatocellular carcinoma. Exp Cell Res 2022;420:113335. [Crossref] [PubMed]
- Chen ZH, Cui MY, Zhang HM. EMX2OS Delays Wilms'Tumor Progression via Targeting miR-654-3p. Ann Clin Lab Sci 2022;52:12-20. [PubMed]
(English Language Editor: J. Gray)


