
Increased phosphorylated CREB1 protein correlates with poor prognosis in clear cell renal cell carcinoma
Introduction
Renal cancer is one of the most prevalent malignancies of the urinary system and approximately 60% to 70% of cases are pathologically identified as the most prevalent subtype-clear cell renal cell carcinoma (ccRCC) (1,2). Compared with that of chromophobe and papillary RCC, the prognosis of ccRCC is relatively unfavorable (3). The identification of novel biomarkers will contribute to effective targeted drugs development and clinical outcomes prediction of ccRCC patients.
Generally, cyclic adenosine monophosphate (cAMP) responsive element binding protein 1 (CREB1) is activated by cAMP, calcium, growth factors, and hormones via multiple signaling pathways (4,5). Once CREB1 is phosphorylated, it upregulates the expression of proto-oncogenes, such as cyclin A and Bcl-2 (6-8), which are associated with cell differentiation and proliferation, the cell cycle, apoptosis, neovascularization, the inflammatory response and tumorigenesis via the ERK1/2, PKA, PKC or CaMKII signaling pathway (5,9). A previous study showed that activated CREB1 became phosphorylated CREB1 at the Ser133 residue (p-CREB1), which bound to the promoter region of downstream genes, including conserved cAMP-responsive elements, and then regulated tumor invasion and proliferation (5). Generally, CREB1 acts as a carcinogenic transcription factor (10), that is overexpressed in many cancers, such as lung cancer (11), mammary carcinoma (12), neuroglioma (13) and gastric cancer (14). Additionally, these studies identified that unfavorable outcomes including tumor recurrence, metastasis and death were correlated with a high level of p-CREB1 protein (14-16).
Here, we evaluated the level of p-CREB1 protein in ccRCC, and identified the correlation of p-CREB1 staining intensity and clinical variables. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/tau-21-371).
Methods
Patients and samples
After acquiring approval from the Institutional Review Board of the Peking University First Hospital (No. 2015-977), a retrospective study was performed on 233 ccRCC patients who underwent partial nephrectomy or radical nephrectomy from January 1, 2004, to December 31, 2010. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all individual participants and patients’ anonymities were preserved. All samples were evaluated by a senior pathologist (QH). Fuhrman nuclear grading was assessed based on the guidelines of Fuhrman et al. (17), and tumor stage was assessed based on the 2010 TNM classification system (18). Tumor metastasis (lymph node metastasis, distant tissue and organ metastasis) and recurrence (relapse in situ or remnant kidney) were confirmed according to radiographic results. Cancer-specific deaths were confirmed by consulting the patients’ immediate family.
Immunohistochemistry of p-CREB1
After the unilateral kidney or renal tumor were surgically removed, the tissues of kidney were fixed by formalin, and then the paraffin embedded samples were cut into 4 µm sections, attached on the slides and subject to immunohistochemical (IHC) staining. After removing the wax, the tissue slides were rehydrated, cultivated for 20 min in a 3% peroxyl aqueous solution, boiled in the EDTA antigen repair solution, and blocked nonspecific proteins in 10% sheep seralbumin for half an hour. Anti-CREB1 (phospho S133) antibody (1:10,000, ab32096; Cambridge, MA, USA) incubated slides about 12 hours at four degrees Celsius. After the slides were lavaged by PBS solution, the slides were conducted with the general IHC kit (PV-6000, ZSGB-BIO, Beijing, China), then dyed with a DAB kit (ZLI-9018, ZSGB-BIO). Some sample slices were included to be counterstained with haematoxylin. Moreover, the primary antibody was superseded with PBS as the negative control.
Interpretation of immunohistochemistry
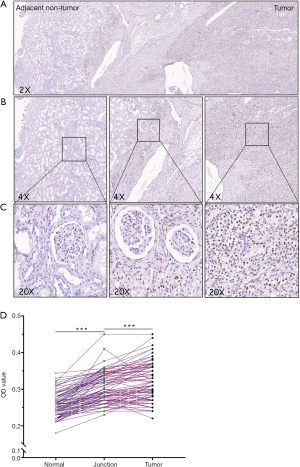
IHC staining was evaluated by a senior pathologist (QH) who was blinded to the patients’ clinical variables. Three or more high-power fields (20×) per tissue section were equally captured by a Leica DMRXA microphotographic system (Leica Biosystems, Germany), and the average staining intensity (optical density, OD) of the cell nucleus was analyzed by a Leica Qwin Standard V2.6 system after normalizing the OD based on the background density of each tissue section. According to the hematoxylin-eosin staining, the tumor-adjacent normal renal tissues, including glomerulus, Bowman’s capsules and kidney tubules, were recognized by pathologist. Following the analysis method above, the staining intensity of the cell nuclei in the tumor-adjacent normal renal cortex, the junction of normal renal tissue and tumor lesion in spite of cell types, and tumor cells was assessed.
Statistical analysis
The variables of different groups were compared using the Chi-square test or nonparametric test, as indicated. Kaplan-Meier survival analysis with Log-rank test to assess the correlation of p-CREB1 classifier and overall, cancer-specific, metastasis-free, or progression-free survival. The univariate Cox regression analysis was used to identify the correlation of variables and survival data, and the variables with a statistic difference (P<0.05) were performed with multivariate Cox regression analysis with forward LR method. All the statistical analyses were carried out by IBM SPSS Statistics 20.0 (IBM SPSS, Chicago, IL) and GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA, USA). A 2-tailed P<0.05 was regarded as statistical difference.
Results
p-CREB1 staining in ccRCC tissues
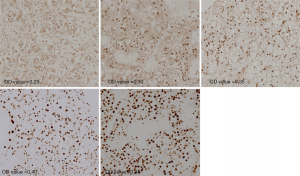
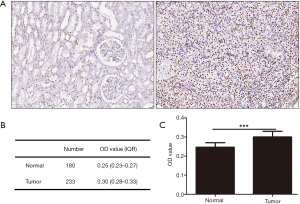
The staining images of p-CREB1 in 233 tumor tissues and paired 180 normal tissues were captured. Among 233 patients, 92 cases were performed IHC staining and the OD of the tumor-adjacent tumor section, the junction region of tumor and normal section and normal section of them were evaluated, respectively. The OD of each picture was computed by Leica Qwin Standard V2.6 software. Microscopic images of different staining intensities with ODs from 0.25 to 0.40 are showed in Figure 1. The staining intensity was significantly increased in tumor sections compared to para-carcinoma tissue sections (P<0.001) (Figure 2), and we found that the staining of p-CREB1 gradually increased from normal tissue to tumor sections (P<0.001) (Figure 3).
Relationship of p-CREB1 staining and clinicopathological features
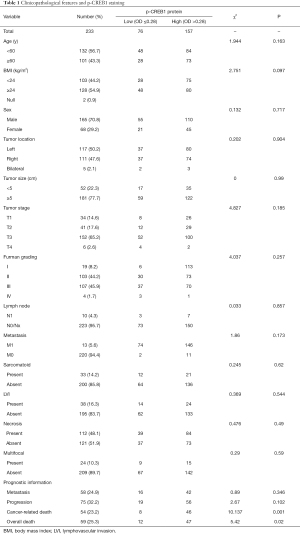
There were 166 (71.2%) males. The median age was 57 years. The median follow-up period was 65 months (range, 3–94 months). During the follow-up, 59 (25.3%) patients died, of which 54 (23.2%) died of renal cancer, 58 (24.9%) had distant metastasis, and 75 (32.2%) had tumor relapse.
On the basis of the receiver operating characteristic curve, all patients were classified into low p-CREB1 staining (OD ≤0.28) and high p-CREB1 staining subgroups (OD >0.28) according to p-CREB1 staining intensity of tumor cells. A chi-square test showed that p-CREB1 staining intensity was associated with cancer-specific mortality (P=0.001) and total mortality (P=0.020), but was not related to age, sex, body mass index, lymph node metastasis, distant metastasis, tumor size or pathological stage classification (Table 1).
Full table
Association of p-CREB1 staining and clinical outcomes
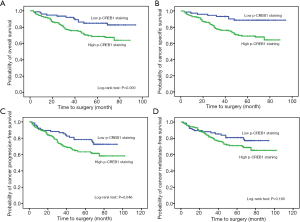
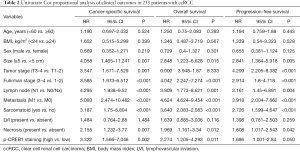
The Kaplan-Meier plot revealed that high p-CREB1staining subgroup was significantly related to poor overall, cancer-specific, and progression-free survival, but there was no significant difference in p-CREB1 classification and metastasis-free survival (Figure 4). In addition, univariate Cox proportional regression analyses demonstrated that high p-CREB1 staining was significantly associated with poor cancer-specific survival and overall survival (Table 2). Multivariate analyses showed that increased p-CREB1 staining was an independent risk factor for cancer specific-free survival [hazard ratio (HR) =4.593, 95% CI: 2.125–9.924, P<0.001], overall survival (HR =3.131, 95% CI: 1.635–5.996, P=0.001) and progression-free survival (HR =2.133, 95% CI: 1.255–3.625, P=0.005) (Table 3).
Full table

Full table
Discussion
In this study, we found that the level of p-CREB1 protein was higher in tumor cells than in normal renal cells and related to poor outcomes for ccRCC patients. Our results verified that CREB1 was an oncogene that led to a poor prognosis for renal carcinoma patients. Li et al. (10) reported that CREB1 was overexpressed in renal tumor cell lines and ccRCC tumor tissues, and further research confirmed that miR-10b-5p and miR-363-3p could directly bind to the mRNA of CREB1, inhibit the protein expression of CREB1 and finally contribute to tumor proliferation and migration. A study launched by Friedrich et al. (19) revealed that CREB1 upregulation in tumors was closely associated with malignant phenotypes, such as tumor stage, grade and LVI. They found miR-22-3p, miR-26a-5p, miR-27a-3p, and miR-221-3p probably inversely regulate CREB1 translation. Huang et al. (20) demonstrated that CREB1 upregulation led to resistance to sorafenib treatment and that red ginseng extract enhanced the anticancer effects of sorafenib by inhibiting the expression of CREB1 in renal cancer (21).
Several studies have found abnormal expression of CREB1 in various carcinomas (10,16,22). Chhabra et al. (16) reported that CREB1 was overexpressed in metastatic breast cancer and promoted tumor cell progression and metastasis. In addition, its upregulation in glioma cells reinforced the transcription of carcinogen microRNA-23a and further enhanced glioblastoma cell proliferation and invasion (23). Shankar et al. (8) reported that CREB1 overexpression was primarily related to the amplification of the CREB1 gene copy number in tumor cells. A study of non-small cell lung cancer showed that the CIP2A positively mediated CREB phosphorylation and promoted cancer progression via cell metabolism (24). In addition, CD44, a marker of cancer stem-like cells, binds to CREB1, promotes CREB1 phosphorylation and further enhances CREB1 recruitment to the cyclin D1 promoter which increases gene transcription, leading to cell proliferation (25).
Generally, the downstream pathways were activated mostly when the CREB1 protein was phosphorylated, so we detected the association of the p-CREB1 protein level and clinical variables. The current study revealed that p-CREB1, independently related to clinical outcomes, was a predictive biomarker of unfavorable prognosis for ccRCC patients. Consistently, the predictive value of CREB1 was identified in a variety of cancers. A study by Seo et al. (15) showed that increased p-CREB1 protein was significantly correlated with increased overall mortality. Wang et al. (14) reported that CREB1, associated with a high death hazard, could act as an independent predictive indicator for gastric cancer patients. Moreover, Chhabra et al. (16) found that a high level of CREB1 transcripts was related to an increased mortality in breast carcinoma. A study performed by Yu et al. (26) demonstrated that CREB1 overexpression was significantly correlated with cancer progression and that the high level of p-CREB1 protein was an independent unfavorable prognostic factor for hepatocellular carcinoma patients. The significant correlation of p-CREB1 and clinical outcomes across demonstrated that p-CREB1 played a crucial role in the process of tumor invasion and metastasis.
One of the limitations of this study is the retrospective nature and we cannot eliminate selection bias from single-center data. Meanwhile, more ccRCC cases need to be included to confirm our results. In addition, in vitro and in vivo studies are required to explore the potential molecular mechanism.
In summary, we estimated p-CREB1 staining by an immunohistochemical method and explored the correlation of its staining intensity and clinicopathological features. Our data demonstrated that p-CREB1 protein was higher in tumor tissues than normal renal cells and could independently predict clinical outcomes. Therefore, our results showed that p-CREB1 protein staining was an effective prognostic factor for ccRCC patients. Targeting p-CREB1, such as by using CREB1 phosphorylation inhibitors, may be a promising therapeutic tactic for this disease.
Conclusions
Our study demonstrated that the level of p-CREB1 protein was higher in tumor tissues than normal renal cells and the staining intensity of p-CREB1 protein was an independent risk factor for the ccRCC patients. Our findings indicated that p-CREB1 is a valuable biomarker and might be a promising therapeutic target for this disease.
Acknowledgments
Funding: This work was supported by grants from the National Key R&D Program of China (2016YFC0902601).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/tau-21-371
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tau-21-371
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tau-21-371). XL and LZ serves as an unpaid editorial board member of Translational Andrology and Urology. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of the Peking University First Hospital (No. 2015-977). Informed consent was taken from all individual participants and patients’ anonymities were preserved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. J Clin Oncol 2014;32:1968-76. [Crossref] [PubMed]
- Beck SD, Patel MI, Snyder ME, et al. Effect of papillary and chromophobe cell type on disease-free survival after nephrectomy for renal cell carcinoma. Ann Surg Oncol 2004;11:71-7. [Crossref] [PubMed]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2001;2:599-609. [Crossref] [PubMed]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 1999;68:821-61. [Crossref] [PubMed]
- Desdouets C, Matesic G, Molina CA, et al. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factors CREB and CREM. Mol Cell Biol 1995;15:3301-9. [Crossref] [PubMed]
- Xiang H, Wang J, Boxer LM. Role of the cyclic AMP response element in the bcl-2 promoter in the regulation of endogenous Bcl-2 expression and apoptosis in murine B cells. Mol Cell Biol 2006;26:8599-606. [Crossref] [PubMed]
- Shankar DB, Cheng JC, Kinjo K, et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell 2005;7:351-62. [Crossref] [PubMed]
- Puri P, Schaefer CM, Bushnell D, et al. Ectopic Phosphorylated Creb Marks Dedifferentiated Proximal Tubules in Cystic Kidney Disease. Am J Pathol 2018;188:84-94. [Crossref] [PubMed]
- Li Y, Chen D, Li Y, et al. Oncogenic cAMP responsive element binding protein 1 is overexpressed upon loss of tumor suppressive miR-10b-5p and miR-363-3p in renal cancer. Oncol Rep 2016;35:1967-78. [Crossref] [PubMed]
- Aggarwal S, Kim SW, Ryu SH, et al. Growth suppression of lung cancer cells by targeting cyclic AMP response element-binding protein. Cancer Res 2008;68:981-8. [Crossref] [PubMed]
- Son J, Lee JH, Kim HN, et al. cAMP-response-element-binding protein positively regulates breast cancer metastasis and subsequent bone destruction. Biochem Biophys Res Commun 2010;398:309-14. [Crossref] [PubMed]
- Cuevas P, Diaz-González D, Carceller F, et al. Dual blockade of mitogen-activated protein kinases ERK-1 (p42) and ERK-2 (p44) and cyclic AMP response element binding protein (CREB) by neomycin inhibits glioma cell proliferation. Neurol Res 2003;25:13-6. [Crossref] [PubMed]
- Wang YW, Chen X, Gao JW, et al. High expression of cAMP-responsive element-binding protein 1 (CREB1) is associated with metastasis, tumor stage and poor outcome in gastric cancer. Oncotarget 2015;6:10646-57. [Crossref] [PubMed]
- Seo HS, Liu DD, Bekele BN, et al. Cyclic AMP response element-binding protein overexpression: a feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer Res 2008;68:6065-73. [Crossref] [PubMed]
- Chhabra A, Fernando H, Watkins G, et al. Expression of transcription factor CREB1 in human breast cancer and its correlation with prognosis. Oncol Rep 2007;18:953-8. [Crossref] [PubMed]
- Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol 1982;6:655-63. [Crossref] [PubMed]
- Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471-4.
- Friedrich M, Heimer N, Stoehr C, et al. CREB1 is affected by the microRNAs miR-22-3p, miR-26a-5p, miR-27a-3p, and miR-221-3p and correlates with adverse clinicopathological features in renal cell carcinoma. Sci Rep 2020;10:6499. [Crossref] [PubMed]
- Huang S, Cui P, Lin S, et al. The Transcription Factor Creb is Involved in Sorafenib-Inhibited Renal Cancer Cell Proliferation, Migration and Invasion. Acta Pharm 2018;68:497-506. [Crossref] [PubMed]
- Kim C, Lee JH, Baek SH, et al. Korean Red Ginseng Extract Enhances the Anticancer Effects of Sorafenib through Abrogation of CREB and c-Jun Activation in Renal Cell Carcinoma. Phytother Res 2017;31:1078-89. [Crossref] [PubMed]
- Shankar DB, Sakamoto KM. The role of cyclic-AMP binding protein (CREB) in leukemia cell proliferation and acute leukemias. Leuk Lymphoma 2004;45:265-70. [Crossref] [PubMed]
- Perry C, Sklan EH, Soreq H. CREB regulates AChE-R-induced proliferation of human glioblastoma cells. Neoplasia 2004;6:279-86. [Crossref] [PubMed]
- Peng B, Lei N, Chai Y, et al. CIP2A regulates cancer metabolism and CREB phosphorylation in non-small cell lung cancer. Mol Biosyst 2015;11:105-14. [Crossref] [PubMed]
- De Falco V, Tamburrino A, Ventre S, et al. CD44 proteolysis increases CREB phosphorylation and sustains proliferation of thyroid cancer cells. Cancer Res 2012;72:1449-58. [Crossref] [PubMed]
- Yu L, Guo X, Zhang P, et al. Cyclic adenosine monophosphate-responsive element-binding protein activation predicts an unfavorable prognosis in patients with hepatocellular carcinoma. Onco Targets Ther 2014;7:873-9. [PubMed]


