
Long ncRNA MALAT1 promotes cell proliferation, migration, and invasion in prostate cancer via sponging miR-145
Introduction
Prostate cancer (PCa) is the most common male cancer. It affected the reproductive system and is considered a serious threat to male health. Since 1975, the rates of PCa have surpassed lung cancer to become the leading male cancer in the United States (1). In 2018, new cases of PCa were expected to cross 1.2 million worldwide (2). The early onset of PCa is asymptomatic, but the symptoms of compression and metastasis become apparent as the disease progresses. Patients usually present at the hospital with complaints of dysuria or hematuria (3). Like other malignancies, PCa requires early diagnosis for better outcomes; once PCa has reached an advanced stage, the 5-year survival rate is only 28% (4). Several theories have been suggested to explain the etiology of PCa in the search for targeted therapy. In addition to surgical options, androgen deprivation therapy (ADT) is the most commonly used treatment. However, after ADT treatment, most patients face the risk of clinical recurrence, and the tumor eventually evolves into castration-resistant prostate cancer (CRPC), which is currently the main cause of death from PCa (5). The clinical results of current anti-cancer treatments are not lasting. Prostate cancer gene 3 (PCA3) is a prostate-specific long non-coding (lncRNA), and as a specific humoral biomarker of prostate cancer, it has been extensively studied in body fluids (6). Therefore, exploring possible therapeutic targets in lncRNAs has broad prospects for the management of PCa patients.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA (>200 nts) that does not encode for a protein. It is also known as nuclear-enriched abundant transcript 2 (NEAT2) and is highly conserved and one of the most abundant lncRNAs in normal tissues (7). The lncRNA MALAT1 was initially reported as a prognostic marker of early-stage non-small cell lung cancer (NSCLC) (8). However, later studies found that MALAT1 is also highly expressed in several other cancers such as lung cancer, hepatocellular cell carcinoma, bladder cancer, colorectal carcinoma, and prostate cancer (9-13). Several studies have now suggested that MALAT1 can promote cell proliferation, migration, invasion, and metastasis in diverse cancers (14-16). In prostate cancer, MALAT1 is positively correlated with Gleason score, prostate-specific antigen (PSA) level, tumor stage, and castration resistance. Loss of MALAT1 expression can induce cell apoptosis (13). In a set of CRPC bone marrow biopsy specimens, MALAT1 was found to be the most highly expressed transcript in CRPC through whole transcriptome sequencing (17). However, the underlying molecular mechanism of MALAT1 in prostate cancer progression has not been fully elucidated.
MicroRNAs (miRNAs) are small non-coding RNAs that contain approximately 23 nucleotides. These play important roles in post-transcriptional repression by base-pairing with the target messenger RNAs (mRNAs) (18). It has been reported that the miRNA miR-145 binds to MALAT1 and regulates its function in cervical cancer (19). Also, miR-145 has been shown to have the ability to down-regulate the expression of SMAD3 and TGFBR2 by directly targeting their 3 prime untranslated regions (3’-UTR) (20-22). The up-regulation of miR-145 and down-regulation of SMAD3 and TGFBR2 is known to inhibit transforming growth factor-β1 (TGF-β1)-induced epithelial-mesenchymal transition (EMT) in bladder cancer (23).
Interestingly, EMT is also known to play a critical role in the cancer metastasis and invasion of prostate cancer (24,25). Therefore, in this study, we aimed to investigate prostate cancer progression mechanism that could be mediated via MALAT1, miR-145, and TGF-β1-induced EMT.
We present the following article in accordance with the ARRIVE Guidelines Checklist (available at http://dx.doi.org/10.21037/tau-20-1526).
Methods
Ethical statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (NO.: KY2020K061) granted by The Second Hospital of Tianjin Medical University, in compliance with national guidelines for the care and use of animals.
Research resource identifiers (RRID)
Antibodies: (Thermo Fisher Scientific Cat# 51-3249-82, RRID:AB_1210532); (US Biological Cat# C0107-03A, RRID:AB_2077545); (Leica Biosystems Cat# NCL-VIM-V9, RRID:AB_442141); (Fitzgerald Industries International Cat# 61R-CD44gMSPE, RRID:AB_1283433); (US Biological Cat# O5403-01B, RRID:AB_2167719); (DSHB Cat# DSHB-hGAPDH-4B7, RRID:AB_2617427); (Miltenyi Biotec Cat# 130-125-235, RRID:AB_2857760); (DSHB Cat# PCRP-SMAD3-1A2, RRID:AB_2619090); (Abbiotec Cat# 250880, RRID:AB_2202462).
Cell Lines: (CLS Cat# 300265/p761_LNCaP, RRID:CVCL_0395); (KCB Cat# KCB 200731YJ, RRID:CVCL_1045).
Software tools: (TargetScan, RRID:SCR_010845); (GraphPad Prism, RRID:SCR_002798); (Starbase V2.0, RRID:SCR_016303).
Study procedures
We first observed the effects of MALAT1 on the proliferation, invasion, metastasis, and EMT process of PCa cells. Then, we examined the relationship among MALAT1, miR-145-5p, and TGFβR2/SMAD3, which are the key downstream factors in the EMT process. Finally, animal experiments were conducted to observe the role of MALAT1 in PCa.
Cell lines, cell culture, and reagents
Human PCa cell lines LNCaP and CWR22Rv1 and normal prostate cells were obtained from the Tianjin Institute of Urology (Tianjin, China). These were maintained in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Gibco, Gaithersburg, MD, USA) with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin antibiotic solution at 37 °C in 5% CO2. Purified recombinant human TGF-β1 protein was purchased from R&D Systems (Minneapolis, MN, USA).
Bioinformatics
Through the gene differential expression analysis between normal prostate tissue and prostate cancer tissue in The Cancer Genome Atlas (TCGA) database, the expression of LncRNA-MALAT1 in PCa tissue was found and confirmed. The absolute value of log2(Fold change) >1 represented differential expression of genes. The greater the value of −log10(pval), the greater the gene expression difference; a P value <0.05 indicated that the difference in MALAT1 expression in different tissue samples was statistically significant.
In various cellular activities, miRNA mediates the occurrence of various regulatory mechanisms by binding to various RNA molecules. The TargetScan (www.targetscan.org/vert_72/) and starBase databases (starbase.sysu.edu.cn) provide information on the interaction between miRNA and various RNA molecules. Among them, miR-145 and MALAT1, miR-145 and SMAD3, and miR-145 and TGFBR2 have targeting relationships, and their potential binding sites can also be found in the database.
Cell transfection
The short hairpin RNAs (shRNAs) sh-MALAT1 and negative control (NC) shRNA were obtained from HanBio (Shanghai, China). The miR-145-5p mimics and mimics NC were acquired from Thermo Fisher Scientific (Waltham, MA, USA). These were transfected into PCa cells using Lipofectamine-3000 as per the manufacturer's protocol.
EMT cell model
The LNCaP and CWR22Rv1 cells were trypsinized and inoculated in 6-well plates, followed by incubation for 24 h.Then, the cells were treated in RPMI-1640 medium containing 2.5% FBS with 5 ng/mL TGF-β1 (Sigma-Aldrich, St. Louis, MO, USA) or a control 5 ng/mL bovine serum albumin (BSA) protein for 2 days. The EMT cell model was transfected with miR-145-5p mimics, mimics NC, sh-MALAT1, or shRNA NC as per the experimental design.
Cell proliferation assay
The viability of PCa cells was examined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. The cells (1×104 cells per well, 100 µL cell suspension) were inoculated in 96-well plates using RPMI-1640 medium. After 24 h incubation, the MTT agent (10 µL of 5 mg/mL) was added to the plates. Then, the cells were incubated for another 4 h, the culture medium was removed, and dimethyl sulfoxide (DMSO, 100 µL/well) was added. The samples’ absorbance was measured at 490 nm using a microplate spectrophotometer (Thermo Fisher Scientific).
Cell migration and invasion assay
The cell migration and invasion were assessed by the wound healing and Transwell invasion assays, respectively. To study cell migration, PCa cells were inoculated in 6-well plates and incubated in the culture medium with 10% FBS until the cells reached 90% confluence. A sterile pipette tip (100 µL) was used to make the scratch through the cell monolayer. After removing the medium, the cells were further cultured in a serum-free medium at 37 °C, 5% CO2, for another 24 h. The images were recorded using a light microscope (Olympus Corporation, Tokyo, Japan) at 100× magnification.
For invasion assay, 24-well Transwell plates (Corning, NY, USA) with Matrigel matrix coating and polycarbonate membrane filter inserts (8 µm pore size, BD Biosciences, San Jose, CA, USA) were used. The PCa cells suspended in a serum-free medium were inoculated in the upper Transwell chamber. The lower Transwell chamber was filled with a culture medium containing 10% FBS. Then, the cells were incubated for another 24 h at 37 °C. The cells that invaded the lower chamber were fixed using 4% paraformaldehyde at room temperature (RT) for 30 min. These were then stained with 0.1% crystal violet for 20 min. The samples were visualized using a light microscope (Olympus Corporation) at 200× magnification.
RNA isolation and qRT-PCR
The PCa cells were lysed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) to extract the total RNA. A QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany) was used to create polymerase chain reaction (PCR) templates and the corresponding complementary DNAs (cDNAs). An ABI Prism 7500 fast real-time PCR system (Applied Biosystems, Forster City, CA, USA) was used to perform the PCR reaction supplemented with cDNAs, primers, and 1× SYBR Master Mix. The relative molecular expression was calculated using the 2−ΔΔCT method, and the normalization was carried out using β-actin as an internal control.
Protein extraction and western blot
On ice lysis of PCa cells was carried out using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China). The total protein sample was obtained by centrifugation at 13,000 ×g for 10 min at 4 °C. After the bicinchoninic acid (BCA) quantification, 20 µg of equal protein samples were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). These were then transferred onto polyvinylidene fluoride (PVDF) membranes. Blocking was carried out using 5% non-fat milk for 1 h, and incubation with the primary antibodies was performed overnight at 4 °C. After washing with Tris-buffered saline (TBS) buffer containing 0.05% Tween-20, the membranes were incubated with secondary antibodies [Anti-rabbit IgG, horseradish peroxidase (HRP)-linked Antibody, #7074, 1/1,000] at RT for 1 h. Protein bands were visualized using a chemiluminescence detection kit (Aidlab Biotechnology, Beijing, China), and intensity was measured using the ImageJ software (National Institutes of Health, Bethesda, MD, USA). Glyceraldehyde 3-phosphate dehydrogenase (GADPH) was used as the internal control. The primary antibodies, anti-E-cadherin (#3195, dilution 1/1,000), anti-N-cadherin (#13116, 1/1,000), anti-Vimentin (#5741, 1/1,000), anti-CD44 (#37259, 1/1,000), anti-Oct-4 (#2750, 1/1,000), anti-SMAD3 (#9523, 1/1,000) and the secondary antibodies were purchased from Cell Signaling Technology (CST, Danvers, MA, USA). Anti-TGF-β1 (ab179695, 1/1,000), anti-TGFBR2 (ab186838, 1/1,000), and anti-GAPDH (ab181602, 1/10,000) primary antibodies were obtained from Abcam (Cambridge, UK).
Luciferase reporter assay
The starBase database (http://starbase.sysu.edu.cn/index.php) and TargetScan database (http://www.targetscan.org/vert_72/) were used to predict the binding sites of miR-145-5p with MALAT1, SMAD3, or TGFBR2. These target binding sites were further verified using a luciferase reporter assay. The MALAT1, SMAD3, or TGFBR2 fragments containing the respective binding sites were amplified and cloned into pMIR-REPORT Luciferase vectors (Promega, Madison, WI, USA) to obtain the MALAT1-WT, SMAD3-WT, or TGFBR2-WT reporter vectors. Similarly, the MALAT1, SMAD3, or TGFBR2 fragments containing the mutant binding sites were also cloned into pMIR-REPORT Luciferase vectors and named as MALAT1-MUT, SMAD3-MUT, or TGFBR2-MUT reporter vectors. As per the experimental design, these vectors were co-transfected with miR-145-5p mimic or mimic NC into the PCa cells using Lipofectamine-3000, followed by incubation for 48 h. A Dual-Luciferase® 1000 Assay kit (Promega) was used to detect the firefly luciferase activity that was normalized to control Renilla luciferase.
RNA-binding protein immunoprecipitation
The PCa cell lysate was centrifuged for 30 min at 12,000 g at 4 °C, and then clear lysates were incubated with the anti-human Ago2 or anti-mouse immunoglobulin G (IgG)-conjugated magnetic beads in an RNA-binding protein immunoprecipitation (RIP) buffer at 4 °C for 4 h. The precipitated RNA from the beads was extracted using TRIzol reagent, and quantity was determined using quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
Subcutaneous tumor formation in nude mice
The experiments were performed under a project license (KY2020K061) granted by The Second Hospital of Tianjin Medical University, in compliance with national guidelines for the care and use of animals. A total of 20 male BALB/C nude mice aged 6 weeks (weight: 18–22 g) were obtained from the Laboratory of Animal Resources, Chinese Academy of Sciences (Beijing, China). These mice were randomly divided into 2 groups of 10 mice each. These were housed in a pathogen-free environment with 50–60% humidity at 21–23 °C under a 12 h light/dark cycle and had free access to food and water. The PCa cells (1×107 cells/mL) suspended in PBS were subcutaneously injected into the right side of the mice’s back ribs. After the tumor’s diameter reached 6–8 mm, the short RNAs, sh-MALAT1/In vivo-jetPEI® (Polyplus-transfection, New York, NY, USA) or shRNA NC/In vivo-jetPEI® were injected into the tumor every 4 days (for a total of 28 days/8 times). The tumor size was measured every 3 days using a Vernier caliper. The volume was calculated using the formula: V = 0.5 L × W2. Here, W and L represent the short diameter and long diameter, respectively. On the 28th day, the mice were sacrificed for further study.
Immunohistochemical assay
The tumor tissue sections (5 µm thick) were made by incision using a microtome (RM2125RT; Leica, Wetzlar, Germany). These were immediately fixed with 4% paraformaldehyde and embedded in paraffin. The standard procedures of Deparaffinization, rehydration, and antigen retrieval were performed. Next, the sections were incubated with the primary antibodies at 4 °C overnight. Then, incubation with secondary antibodies was performed for 30 min at room temperature. After washing with PBS, the sections were stained with diaminobenzidine (DAB, Sigma-Aldrich) and counterstaining was carried out using hematoxylin (Sigma-Aldrich). The pictures were recorded using a light microscope (Olympus BX 51). The primary antibodies Anti-E-cadherin (ab40772, 1/500), anti-N-cadherin (ab18203, 1/1,000), anti-Vimentin (ab92547, 1/500), anti-CD44 (ab157107, 1/1,000), anti-Oct-4 (ab181557, 1/1,000), anti-c-met (ab51067, 1/250), and anti-Nanog (ab109250, 1/250), and secondary antibodies (Goat Anti-rabbit IgG, HRP-linked Antibody, ab205718, 1/2,000) were purchased from Abcam.
Statistical analysis
Each experiment was repeated 3 times. The data were analyzed using the GraphPad Prism 7 software (GraphPad Software, San Diego, CA, USA) and are presented as mean ± SD when measurement data were normally distributed. The student’s t-test assessed the differences between the 2 groups. The difference analysis among multiple groups was performed using one-way analysis of variance (ANOVA) with Bonferroni post hoc comparison test when each group satisfies normality, and variance was homogeneous between groups. In all statistical analyses, all tests were 2-sided, and a P value of less than 0.05 was considered a significant difference.
Results
MALAT1 was overexpressed in PCa tissues and cell lines
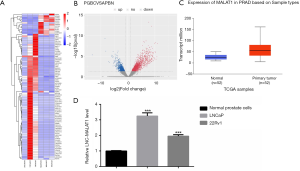
To assess the expression level of MALAT1 in PCa, raw microarray data obtained from TCGA datasets were analyzed using the heat map volcano plot. The differential gene cluster analysis suggested a variation in MALAT1 expression between the PCa tissues and the corresponding adjacent normal tissues (Figure 1A,B). Moreover, TCGA datasets revealed that MALAT1 was highly expressed in PCa tissues compared with non-tumor tissues (Figure 1C). To verify these findings, qRT-PCR experiments were performed using LNCaP and CWR22Rv1 cells. Here too, we found that the MALAT1 expression was upregulated in PCa cell lines compared to normal prostate cells (Figure 1D). Overall, these results suggested that MALAT1 is highly expressed in PCa and could be a tumor-promoting factor.
Suppression of MALAT1 reduced the proliferation, migration, and invasion of the PCa cells
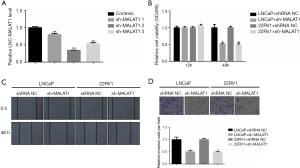
To evaluate the role of MALAT1 over-expression in cell proliferation, migration, and invasion, knockdown of MALAT1 was carried out using the short RNA sh-MALAT1. The qRT-PCR results showed that this knockdown significantly down-regulated the expression of MALAT1 in LNCaP and CWR22RV1 cells (Figure 2A). Furthermore, the MTT assays results also suggested that the down-regulation of MALAT1 significantly inhibited the proliferation capability of LNCaP and CWR22RV1 cells (Figure 2B). Next, we performed wound-healing assays and Transwell assays to find the role of MALAT1 in the migratory and invasive abilities of LNCaP and CWR22RV1 cells. The results from wound-healing assays showed that the suppression of MALAT1 remarkably reduced LNCaP and CWR22RV1 cells (Figure 2C). Similar results were obtained from the Transwell assays. Here too, transfection of sh-MALAT1 weakened the invasive abilities of LNCaP and CWR22RV1 cells (Figure 2D). Taken together, these results suggested that suppression of MALAT1 significantly reduces the proliferation, migration, and invasion abilities of PCa cells.
Knockdown of MALAT1 inhibits TGF-β1-induced EMT in PCa cells
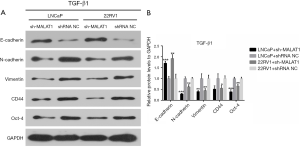
To investigate the relationship between MALAT1 and TGF-β1-induced EMT in PCa cells, the protein levels of epithelial marker E-cadherin, the mesenchymal markers N-cadherin and Vimentin, and cancer stem-like cell markers CD44 and Oct-4 were measured using western blot in TGF-β1-treated cells. The results showed that sh-MALAT1 based knockdown of MALAT1 dramatically increased E-cadherin expression while N-cadherin, Vimentin, CD44, and Oct-4 were suppressed in LNCaP and CWR22RV1 cells (Figure 3). This clearly indicated the role of MALAT1 in TGF-β1-induced EMT and cell dedifferentiation in PCa cells.
MALAT1 directly targets miR-145 in PCa cells
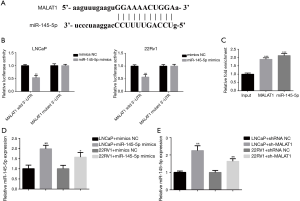
To understand the potential molecular mechanisms by which MALAT1 could regulate tumor progression, we carried out bioinformatics analysis using starBase v3.0. The program predicted the potential binding region between MALAT1 and miR-145-5p, suggesting that miR-145-5p could target MALAT1 (Figure 4A). To verify this, we conducted luciferase reporter assays in PCa cells. The results showed that the co-transfection of miR-145-5p mimics with luciferase reporter plasmid MALAT1-WT led to reduced luciferase activities in LNCaP and CWR22RV1 cells, while no effect was found if it was transfected with MALAT1-MUT plasmid (Figure 4B). We further carried out RIP assays to test whether miR-145 could recruit MALAT1 to RNA-induced silencing complex (RISC) for mRNA degradation. The Ago2 protein is part of minimal RISC. When pull-down experiments were performed, qRT-PCR showed that levels of MALAT1 and miR-145-5p were higher in Ago2 precipitates compared with the input sample (Figure 4C). Furthermore, qRT-PCR showed that the expression level of miR-145-5p was down-regulated (Figure 4D),but knockdown of MALAT1 dramatically increased the expression of miR-145-5p in LNCaP and CWR22RV1 cells (Figure 4E). Overall, these results suggested that MALAT1 directly targeted the miR-145 and functions as a competing endogenous RNA (ceRNA) for miR-145-5p.
MALAT1 regulates SMAD3 and TGFBR2 through competitively binding to miR-145
It was discovered that SMAD3 and TGFBR2 were downstream targets of miR-145. The potential binding regions between them and miR-145 were predicted using TargetScan (Figure 5A,B). These interactions were confirmed experimentally using dual-luciferase reporter assays. The results showed that the transfection of miR-145-5p mimics with WT SMAD3 or WT TGFBR2 resulted in a marked decrease of luciferase activity whereas no obvious change in luciferase activity was noticed transfection with SMAD3 and TGFBR2 mutant plasmids in LNCaP and CWR22RV1 cells (Figure 5C,D). Upon TGF-β1 induction, the levels of SMAD3 and TGFBR2 were upregulated. But when the LNCaP and CWR22RV1 cells were treated with miR-145 mimics, levels of SMAD3 and TGFBR2 decreased significantly (Figure 5E,F). This demonstrated that miR-145 could play an inhibitory role in TGF-β1-induced EMT. Interestingly, when the LNCaP and CWR22RV1 cells were transfected with sh-MALAT1, the expression levels of SMAD3 and TGFBR2 were reduced further (Figure 5G,H). This suggested that miR-145 acts as a protective factor in PCa, and MALAT1 promotes TGF-β1-induced EMT via the miR-145-5p-SMAD3/TGFBR2 axis. It could be concluded that suppression of MALAT1 could down-regulate SMAD3 and TGFBR2 and therefore inhibit TGF-β1-induced EMT via competitively binding to miR-145 in PCa.

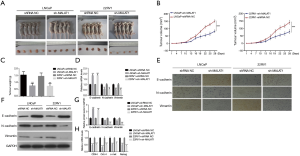
Suppression of MALAT1 inhibits tumorigenesis and EMT in nude mice model
Next, we carried out the in vivo assays to further verify the role of MALAT1 in PCa. The nude mice were subcutaneously inoculated with the sh-MALAT1 or shRNA-NC transfected LNCaP and CWR22RV1 cells (Figure 6A). After inoculation, the tumor tissues’ corresponding volumes were measured up to 28 days, and tumor tissues were isolated on the 28th day (Figure 6A,B). The results indicated that the growth rate of MALAT1-silenced mice tumors was much slower compared with the control group tumors. Also, the weight of tumors isolated on the 28th day from the sh-MALAT1 mice was significantly lower than the tumors isolated from the control (shRNA-NC treated) group (Figure 6C). Furthermore, the results from qRT-PCR, immunohistochemical (IHC) assay and western blot suggested that the epithelial marker E-cadherin was upregulated and the mesenchymal markers N-cadherin and Vimentin were down-regulated in MALAT1-silenced tumor tissues compared with the control group (Figure 6D,E,F,G). The cancer stem-like cell markers CD44, Oct-4, c-met, and Nanog, were down-regulated in sh-MALAT1 treated tumors compared with shRNA-NC treated tumors (Figure 6H). Altogether, the findings of the role of MALAT1 in tumorigenesis and EMT were also consistent in the in vivo mice experiments.
Discussion
As per the Global Burden of Disease 2013 (GBD2013), the number of new cases and deaths of PCa in China has increased drastically (26). To improve the prognosis in PCa patients, it is urgently important to explore applied biomarkers to facilitate early diagnosis and optimize treatment strategies. In recent decades, identifying PSA as a diagnostic biomarker for PCa significantly decreased PCa specific mortality (27). However, PSA can also be elevated in patients with benign prostatic hyperplasia, prostatitis, and other non-malignant diseases. Therefore, using PSA as a routine screening method for prostate cancer is debatable (28,29). Using circulating mRNAs as potential biomarkers for cancer diagnosis and prognosis has recently been a great success (30,31). To date, MALAT1 has been considered a poor candidate for prostate cancer prognosis (13). However, our study's results strongly emphasized that MALAT1 is overexpressed in PCa and should be used as a novel prognostic factor.
We know that MALAT1 is involved in cancers and a variety of other pathophysiological processes such as diabetes (32). Moreover, various molecular mechanisms of MALAT1 for modulating cancer invasion and metastasis have been reported. For instance, Fan et al. showed that MALAT1 binds to SUZ12, a part of the PRC2 complex, to regulate EMT in bladder cancer (11). It has also been reported that MALAT1 functions as ceRNA for several miRNAs such as miR-145 (19), miR-200c (33), miR-202 (34), miR-204 (35), miR-1 (36), and miR-206 (37). Furthermore, MALAT1 is known to regulate numerous signaling pathways such as WNT (38), mTOR (39), PI3K-AKT (40), and MAPK (41) in several cancer types. In this study, we focused on elucidating the molecular mechanism of MALAT1 function as a ceRNA of miR-145 in PCa progression.
It is known that miR-145 inhibits the growth of tumor cells by interacting with multiple target genes. Its expression becomes downregulated in a variety of tumor cells. Essentially, miR-145 acts as a tumor suppressor in several cancer types such as bladder cancer, breast cancer, and colorectal carcinoma (42-44). Here too, we showed that expression of miR-145 was down-regulated in PCa, the progression of which relies heavily on the process of EMT (45,46). We found that miR-145 directly binds to the 3’-UTR of the key downstream target genes SMAD3 and TGFBR2 of the TGF-β signaling pathway and suppresses TGF-β1-induced EMT. The receptor CD44s is known to modulate the TGFβ-mediated mesenchymal phenotypes and has been related to the poor prognosis of hepatocellular carcinoma (47). In PCa, too, CD44 plays a positive role in TGF-β1-induced EMT (48). Therefore, apart from the epithelial marker E-cadherin, and the mesenchymal markers N-cadherin and Vimentin, cancer stem-like cell markers CD44, Oct-4, c-met, and Nanog were also investigated to understand the process of EMT.
Conclusions
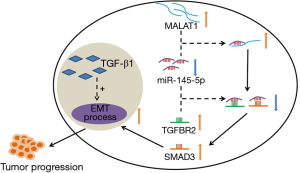
A holistic molecular mechanism of MALAT1 promoting prostate cancer through TGF-β1-induced EMT via miR-145 is depicted in Figure 7. Poor prognosis in PCa patients is closely related to the high expression of MALAT1. The process of TGF-β1-induced EMT is critical for cancer progression, and miR-145 functions as its suppressor. However, MALAT1 directly binds to miR-145 to inhibit its suppressor role. In other words, MALAT1 indirectly promotes TGF-β1-induced EMT by sequestering miR-145. Therefore, we suggest that MALAT1 can be used as a potential prognostic factor and biomarker of PCa.
Acknowledgments
Funding:This study was supported by Tianjin Technical Expert Project (No. 20YDTPJC00370), Fund of the Second Hospital Tianjin Medical University (No. 2020ydey04); Clinical Research Fund of the Second Hospital Tianjin Medical University (No. 2020LC06); The Science & Technology Development Fund of Tianjin Education Commission for Higher Education (No. 2018KJ050); Natural Science Foundation of Tianjin (No. 19JCYBJC26900); Traditional Chinese medicine combined with Western medicine research project (No. 2019137); Science and Technology Talent Cultivation Project of Tianjin Municipal Health Commission (No. KJ20137); and China Postdoctoral Science Foundation Grant (No. 2019M660060).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1526
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1526
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1526). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (NO.: KY2020K061) granted by The Second Hospital of Tianjin Medical University, in compliance with national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Nanda A, Chen MH, Moran BJ, et al. Neoadjuvant hormonal therapy use and the risk of death in men with prostate cancer treated with brachytherapy who have no or at least a single risk factor for coronary artery disease. Eur Urol 2014;65:177-85. [Crossref] [PubMed]
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- Yuan X, Cai C, Chen S, et al. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene 2014;33:2815-25. [Crossref] [PubMed]
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999;59:5975-9. [PubMed]
- Hutchinson JN, Ensminger AW, Clemson CM, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007;8:39. [Crossref] [PubMed]
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41. [Crossref] [PubMed]
- Gutschner T, Hammerle M, Diederichs S. MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791-801. [Crossref] [PubMed]
- Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol 2012;29:1810-6. [Crossref] [PubMed]
- Fan Y, Shen B, Tan M, et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res 2014;20:1531-41. [Crossref] [PubMed]
- Xu C, Yang M, Tian J, et al. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol 2011;39:169-75. [PubMed]
- Ren S, Liu Y, Xu W, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol 2013;190:2278-87. [Crossref] [PubMed]
- Li L, Feng T, Lian Y, et al. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A 2009;106:12956-61. [Crossref] [PubMed]
- Ji Q, Zhang L, Liu X, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer 2014;111:736-48. [Crossref] [PubMed]
- Bi S, Wang C, Li Y, et al. LncRNA-MALAT1-mediated Axl promotes cell invasion and migration in human neuroblastoma. Tumour Biol 2017;39:1010428317699796 [Crossref] [PubMed]
- Sowalsky AG, Xia Z, Wang L, et al. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Cancer Res 2015;13:98-106. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Lu H, He Y, Lin L, et al. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol 2016;37:1683-91. [Crossref] [PubMed]
- Megiorni F, Cialfi S, Cimino G, et al. Elevated levels of miR-145 correlate with SMAD3 down-regulation in cystic fibrosis patients. J Cyst Fibros 2013;12:797-802. [Crossref] [PubMed]
- Huang H, Sun P, Lei Z, et al. miR-145 inhibits invasion and metastasis by directly targeting Smad3 in nasopharyngeal cancer. Tumour Biol 2015;36:4123-31. [Crossref] [PubMed]
- Hu J, Qiu M, Jiang F, et al. MiR-145 regulates cancer stem-like properties and epithelial-to-mesenchymal transition in lung adenocarcinoma-initiating cells. Tumour Biol 2014;35:8953-61. [Crossref] [PubMed]
- Liu X, Wu Y, Zhou Z, et al. Celecoxib inhibits the epithelial-to-mesenchymal transition in bladder cancer via the miRNA-145/TGFBR2/Smad3 axis. Int J Mol Med 2019;44:683-93. [Crossref] [PubMed]
- Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004;117:927-39. [Crossref] [PubMed]
- Khan MI, Adhami VM, Lall RK, et al. YB-1 expression promotes epithelial-to-mesenchymal transition in prostate cancer that is inhibited by a small molecule fisetin. Oncotarget 2014;5:2462-74. [Crossref] [PubMed]
- Qi JL, Wang LJ, Zhou MG, et al. Disease burden of prostate cancer among men in China, from 1990 to 2013. Zhonghua Liu Xing Bing Xue Za Zhi 2016;37:778-82. [PubMed]
- Stamey TA, Caldwell M, McNeal JE, et al. The prostate specific antigen era in the United States is over for prostate cancer: what happened in the last 20 years? J Urol 2004;172:1297-301. [Crossref] [PubMed]
- Xiao LH, Chen PR, Gou ZP, et al. Prostate cancer prediction using the random forest algorithm that takes into account transrectal ultrasound findings, age, and serum levels of prostate-specific antigen. Asian J Androl 2017;19:586-90. [Crossref] [PubMed]
- Armstrong BK, Barry MJ, Frydenberg M, et al. PSA testing for men at average risk of prostate cancer. Public Health Res Pract 2017;27:2731721 [Crossref] [PubMed]
- Gahan PB, Swaminathan R. Circulating nucleic acids in plasma and serum. Recent developments. Ann N Y Acad Sci 2008;1137:1-6. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Wu Y, Huang C, Meng X, et al. Long Noncoding RNA MALAT1: Insights into its Biogenesis and Implications in Human Disease. Curr Pharm Des 2015;21:5017-28. [Crossref] [PubMed]
- Li Q, Zhang C, Chen R, et al. Disrupting MALAT1/miR-200c sponge decreases invasion and migration in endometrioid endometrial carcinoma. Cancer Lett 2016;383:28-40. [Crossref] [PubMed]
- Zhang Y, Chen Z, Li MJ, et al. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 regulates the expression of Gli2 by miR-202 to strengthen gastric cancer progression. Biomed Pharmacother 2017;85:264-71. [Crossref] [PubMed]
- Hou Z, Xu X, Zhou L, et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol 2017;39:1010428317718135 [Crossref] [PubMed]
- Jin C, Yan B, Lu Q, et al. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol 2016;37:4025-33. [Crossref] [PubMed]
- Wang SH, Zhang WJ, Wu XC, et al. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget 2016;7:37857-67. [Crossref] [PubMed]
- Ji Q, Liu X, Fu X, et al. Resveratrol inhibits invasion and metastasis of colorectal cancer cells via MALAT1 mediated Wnt/beta-catenin signal pathway. PLoS One 2013;8:e78700 [Crossref] [PubMed]
- Malakar P, Shilo A, Mogilevsky A, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res 2017;77:1155-67. [Crossref] [PubMed]
- Jin Y, Feng SJ, Qiu S, et al. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharmacol Sci 2017;21:3176-84. [PubMed]
- Wu XS, Wang XA, Wu WG, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther 2014;15:806-14. [Crossref] [PubMed]
- Ichimi T, Enokida H, Okuno Y, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer 2009;125:345-52. [Crossref] [PubMed]
- Spizzo R, Nicoloso MS, Lupini L, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ 2010;17:246-54. [Crossref] [PubMed]
- Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 2007;72:397-402. [Crossref] [PubMed]
- Matuszak EA, Kyprianou N. Androgen regulation of epithelial-mesenchymal transition in prostate tumorigenesis. Expert Rev Endocrinol Metab 2011;6:469-82. [Crossref] [PubMed]
- Grant CM, Kyprianou N. Epithelial mesenchymal transition (EMT) in prostate growth and tumor progression. Transl Androl Urol 2013;2:202-11. [PubMed]
- Mima K, Okabe H, Ishimoto T, et al. CD44s regulates the TGF-beta-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res 2012;72:3414-23. [Crossref] [PubMed]
- Shang Z, Cai Q, Zhang M, et al. A switch from CD44(+) cell to EMT cell drives the metastasis of prostate cancer. Oncotarget 2015;6:1202-16. [Crossref] [PubMed]


