
Biomarkers for detection of clinically significant prostate cancer: contemporary clinical data and future directions
Introduction
Prostate cancer (PCa) remains one of the most common malignancies and a leading cause of death in men worldwide (1-3). In the United States alone, PCa accounts for more than 20% of all cancer diagnoses in men, with 190,000 cases and upwards of 33,000 PCa deaths projected in 2020 (4). Screening with prostate-specific antigen (PSA) has been shown to reduce mortality in men with clinically significant PCa [Grade Group (GG) ≥2] (5-8) and remains the mainstay of PCa detection, despite well-studied drawbacks. PSA is prostate gland-specific, but not cancer-specific (9,10). As such, widespread PSA screening results in frequent negative prostate biopsy and overdiagnosis of indolent disease (11,12), subjecting patients to undue harms in the course of diagnostic evaluation (13). Meanwhile, traditionally only about one-third of men with elevated PSA are found to have PCa on biopsy, with even fewer harboring GG ≥2 disease (14,15).
The limitations of PSA have led to development of novel biomarkers aimed at better informing the risk of GG ≥2 cancer (16). We herein provide a review of serum and urine biomarkers clinically-available to aid in diagnosis of GG ≥2 PCa, including the Prostate Health Index (PHI), 4-Kallikrein score (4Kscore), SelectMDx, ExoDx Prostate Intelliscore (EPI), and MyProstateScore (MPS) (Table 1).
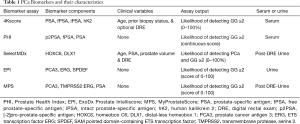
Full table
Study selection
The PubMed database was queried by biomarker name and resulting abstracts were reviewed in March of 2020. Our query included commercially-available serum- and urine-based biomarkers proposed for use following elevated PSA to improve the specificity of screening (17). Of note, preliminary MPS data was available at the time of initial review and formally cited at the manuscript revision stage. The primary outcome of interest was GG ≥2 PCa, and we included post-discovery (i.e., validation) studies that provided sensitivity and specificity for GG ≥2 PCa or provided sufficient raw data for calculation. We included studies of patients referred for prostate biopsy, in the vast majority of cases due to elevated PSA and/or abnormal digital rectal examination (DRE). Because these tests are proposed to aid in the diagnostic evaluation of at-risk men rather than primary screening, we included cohorts with a GG ≥2 prevalence >10%. To provide clinical context, data were stratified by study population [overall (i.e., all patients referred for prostate biopsy) vs. specific clinical criteria (i.e., specific PSA ranges)] and by biopsy status (initial vs. repeat biopsy). Each variable and summary statistic is defined in Table 2.
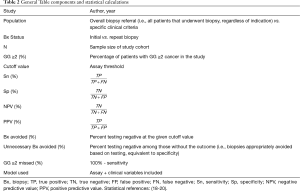
Full table
Measures of diagnostic performance
We reported the sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV) of each biomarker at one or more threshold values [calculated values are labeled with an asterisk (*)]. The purpose of the following subsections is to briefly summarize relevant calculations and interpretation of common statistical parameters (18-20). Although beyond the scope of this review, it is important to consider that these measures are impacted by different study designs (i.e., matched) and potential confounders, requiring recalculation/adjustment for proper interpretation (21,22).
Sensitivity, specificity, biopsies avoided, and GG ≥2 PCa missed
Sensitivity and specificity are common measures of diagnostic accuracy that quantify the agreement of a test relative to a reference standard. Sensitivity and specificity depend on the threshold value used to identify positive (above the threshold) and negative (below the threshold) test results. Changing the threshold to increase sensitivity will necessarily decrease the specificity. In contrast to predictive values, sensitivity and specificity are inherent characteristics of a test and do not vary based on disease prevalence in the study population.
Sensitivity is the proportion of patients that have a positive test among all patients that truly do have the condition (“positivity in disease”); it is also called the true positive (TP) rate (20). In the current context, sensitivity represents the proportion of men that have a positive test among all that truly have GG ≥2 cancer. Specificity represents the proportion of patients with a negative test among all patients that truly do not have the condition (“negativity in health”). In this context, specificity is the proportion of men that have a negative test among all men that do not have GG ≥2 cancer. The false positive rate is equal to 1 – specificity.
As an example, we can apply the use of a biomarker test to determine the need for prostate biopsy: patients with a positive test do undergo biopsy, and patients with a negative test do not. For illustrative purposes, we will imagine that biopsy is 100% accurate for detecting GG ≥2 PCa if it exists. A test with 97% sensitivity would be positive for 97% of men with GG ≥2 cancer, appropriately leading to biopsy for those patients. The test would be negative in 3% of men with GG ≥2 cancer, leading to (inappropriately) not performing biopsy and therefore missing 3% of GG ≥2 cancers. As such, the proportion of GG ≥2 cancers missed with a given testing approach can be calculated as 100%—sensitivity. This underscores the importance of a highly sensitive test when the main clinical aim is to avoid missing diagnoses (for example, when the condition is highly lethal but curable with treatment).
A test with 30% specificity will yield a negative test result in 30% of patients that do not have GG ≥2 cancer. As biopsy will be appropriately avoided in such men, specificity equals the proportion of unnecessary biopsies (i.e., biopsies that would have been negative/GG1) that were avoided through use of the test. This is in contrast to overall “biopsies avoided”, which is simply the percentage of men that have a negative test result, without accounting for whether or not the negative test result accurately reflected the underlying disease state.
Negative and positive predictive values
Unlike sensitivity and specificity, predictive values are dependent on the prevalence of the outcome in the population. Among all patients with a negative test, the NPV is the proportion of patients that truly do not have GG ≥2 disease. For instance, a biomarker with 90% NPV for GG ≥2 PCa indicates that there is a 90% probability of not having GG ≥2 disease in patients who test negative. By contrast, among all men with a positive test the PPV is the proportion of men that do have GG ≥2 disease. If an assay has a PPV of 30% then a positive test indicates a 30% probability of having GG ≥2 PCa. It is additionally assumed for the purpose of this review that the patient/population presenting for biomarker testing is consistent with the population in which assay performance measures were derived. Thus, we have aimed to clearly summarize pertinent clinical data for all studies.
Discrimination
The area under the receiver operating characteristic curve quantifies the ability of a test to discriminate between those with and without the outcome. The AUC measures the area under the ROC curve, which plots the sensitivity versus the false positive rate for all potential threshold values. Importantly, the false-positive rate (1 – specificity) is the probability of a positive test result when the condition is absent. Meanwhile, the false-negative rate (1 – sensitivity) is the probability of a negative test result when the condition is present. For binary outcomes, the AUC is identical to the concordance statistic (c-statistic) (23). While the AUC provides a broad measure of performance across all potential thresholds, it values all potential cutoffs equally, making it limited for interpreting potential clinical utility of a test. For example, a test threshold that fails to detect 75% of high-grade cancers (25% sensitivity) would not be acceptable for clinical use, and therefore the false positive rates that determine the AUC are irrelevant at such low sensitivities. Yet the area under the curve for sensitivity 0–25% contributes the same proportion to the overall AUC as the clinically-meaningful sensitivity range of 75–100%. In the setting of GG ≥2 prostate cancer, useful tests generally have sensitivities of at least 75%, and, ideally, >90%. Therefore, while we present AUC as a broad measure of discrimination, we have focused on the accuracy of tests at specific thresholds presented in the literature. The performance of various testing approaches to support clinical decision-making is best summarized with decision-analytic measures such as decision curve analysis reviewed elsewhere (24).
Biomarkers
The Prostate Health Index (PHI)
The Beckman Coulter PHI is a blood-based assay that combines [-2] proPSA (p2PSA), free PSA (fPSA), and total PSA (tPSA) into a single score to predict likelihood of PCa on biopsy. Initial data revealed that use of PSA isoforms such as percent free PSA (%fPSA = fPSA/tPSA) could improve PCa detection relative to PSA (25), and additional evidence supporting the use of PSA isoforms led to the development of PHI. In a 2011 multicenter study of 892 patients, PHI demonstrated greater AUC (0.70) than its individual components [p2PSA (AUC 0.56), fPSA (AUC 0.62), and tPSA (AUC 0.53)] for PCa in men with PSA 2–10 ng/mL and normal DRE (26). Subsequent studies in the overall referral population (27-29) and in men with PSA 2–10 ng/mL (30-35) demonstrated improvements in AUC ranging from 0.06 to 0.25 relative to PSA-based models.
More relevant to contemporary practice, a number of studies have characterized the use of PHI for predicting GG32 cancer (Table 3).
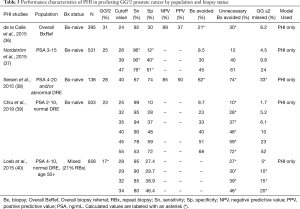
Full table
In 395 men referred for initial biopsy, regardless of PSA level, de la Calle et al. showed that a PHI cutoff of 24 demonstrated 92% sensitivity, 30% specificity, 89% NPV, and 37% PPV for GG ≥2 PCa. Using this cutoff could have avoided 21% of biopsies and 30% of unnecessary biopsies while delaying the diagnosis of 8.2% of GG ≥2 cancers (36).
Several studies have assessed PHI within specific PSA ranges in biopsy-naïve men. Among three such studies, PHI was shown to discriminate GG ≥2 PCa on biopsy with AUC 0.71 (N=503, PSA 2–10 ng/mL and normal DRE) (39), AUC 0.71 (N=531, PSA 3–15 ng/mL) (37), and AUC 0.80 (N=138, PSA 4–20 ng/mL) (38). Improvements in accuracy compared to PSA-based models ranged from 0.08 to 0.13 across these studies. Sensitivities and specificities using various PHI thresholds are listed in Table 3. Notably, Nordström et al. and Seisen et al. found that PHI outperforms base clinical models of PSA and age (AUC 0.71 vs. 0.55) (37) and PSA density (PSAD) (AUC 0.80 vs. 0.68) (38), respectively, thus helping to discriminate between GG1/benign and GG ≥2 tissue.
In 2015, Loeb et al. assessed prospectively collected, multicenter data on 658 men with PSA 4–10 ng/mL and normal DRE, of which 21% had a history of prior negative biopsy. Using a PHI threshold for biopsy of 28.6 led to 90% sensitivity and could’ve avoided 30% of unnecessary biopsies, thus demonstrating a potential role of PHI in further risk stratification of patients meeting these clinical criteria (40). PHI was approved by the Food and Drug Administration (FDA) in 2012 for select men (at least 50 years of age, non-suspicious DRE, and PSA 4–10 ng/mL) (41), and the European Association of Urology (EAU) mention PHI may be offered in a subset of patients (PSA 2–10 ng/mL, non-suspicious DRE) to better define the risk of GG ≥2 cancer (42).
The 4-kallikrein score
The OPKO Health 4-kallikrein score (4Kscore) is a blood-based test consisting of a 4-kallikrein panel [PSA, fPSA, intact PSA (iPSA), and human kallikrein 2 (hK2)] plus age, prior biopsy status, and DRE findings (optional). The 4Kscore is reported as a percent likelihood of harboring GG ≥2 PCa (0–100%). Using data from the European Randomized Study of Screening for Prostate Cancer (ERSPC), four early studies by Vickers and colleagues evaluated a 4Kscore (4K panel, age, and DRE status) threshold of 20% in biopsy-naïve referral populations. These data indicated that 36–60% of biopsies could have been avoided while missing 2.3–12% of GG ≥2 cancers, with AUCs ranging from 0.80 to 0.90 (43-46). Relative to the contemporary population in which biomarker testing is often applied (GG ≥2 PCa prevalence approximating 16–36%) (40,47-51), these cohorts were of lower risk (4.3–9.9% GG ≥2 PCa). This work set the foundation for future 4Kscore validation studies.
In 2015, Parekh and colleagues published a large prospective validation study of 1,012 men referred for biopsy (i.e., overall referral population) across 26 U.S. centers, of which 22% had a history of prior negative biopsy. The authors observed an AUC of 0.82 for predicting GG ≥2 disease, which significantly outperformed a modified Prostate Cancer Prevention Trial Risk Calculator (PCPT-RC) 2.0 clinical model (AUC 0.74). Based on this, use of 4Kscore could have potentially avoided 30–58% of biopsies across different thresholds (6%, 9%, 12%, and 15%) while missing 1.3–4.7% of GG ≥2 cancers (47). These findings are corroborated by more recent studies using the 4Kscore in the overall referral population, which have yielded AUC ranges from 0.81 to 0.83 with improvements in AUC of 0.07 to 0.16 compared to a PSA-based clinical model and a urinary biomarker assay (SelectMDx), respectively (49,51). 4Kscore validation studies aimed at detection of GG ≥2 cancer are illustrated in Table 4.
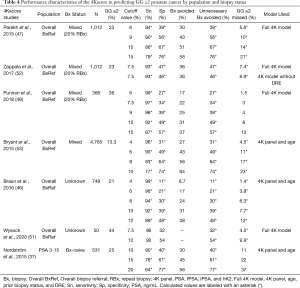
Full table
SelectMDx
The SelectMDx (MDxHealth) urine-based assay incorporates biomarkers homeobox C6 (HOXC6) and distal-less homeobox 1 (DLX1) with clinical factors of age, PSA, prostate volume, and DRE findings to estimate percent likelihood of PCa and percent likelihood of GG ≥2 on biopsy. SelectMDx validation studies aimed at detection of GG ≥2 cancer are summarized in Table 5.

Full table
Early work by Leyten et al. first identified three genes in urinary sediment for the detection of overall and GG ≥2 PCa: HOXC6, DLX1, and Tudor domain-containing protein 1 (TDRD1) (54). A follow-up study by Van Neste and colleagues focused on urinary mRNA levels of two of these genes (HOX6 and DLX1) (55). Unfortunately, the pertinence of these findings to contemporary practice are limited by study cohort characteristics, with mean PSA values greater than 10 ng/mL—well beyond the range in which additional testing is recommended by expert guidelines (17). Nonetheless, a model including HOXC6 and DLX1 was applied to a validation cohort of 386 men. Notably, the multigene model provided a lower AUC (0.86, 95% CI, 0.80–0.92) than a baseline model of clinical variables only (AUC 0.87, 95% CI, 0.81–0.93). When DRE findings were removed from the multigene model, the AUC improved to 0.90 (95% CI, 0.85–0.95). While these data were not particularly promising, they laid the groundwork for additional efforts.
A recent study of biopsy-naïve patients by Haese et al. evaluated SelectMDx in the overall referral population (N=916) and in a subgroup of men with PSA <10 (N=715). The full SelectMDx model consisted of urinary HOXC6 and DLX1 mRNA plus age, PSAD, and DRE result (without incorporating history of prior biopsy). In the overall biopsy-naïve population, SelectMDx demonstrated 93% sensitivity, 47% specificity, and 95% NPV. Similar findings were observed in men with PSA <10 ng/mL, with 89% sensitivity, 53% specificity, and 95% NPV. Notably, when assessed without prostate volume (i.e., PSAD was replaced with PSA), SelectMDx had decreased sensitivity (87%), specificity (38%), and NPV (92%) (50). The inclusion of prostate volume in the SelectMDx model has implications for clinical use, particularly in biopsy-naïve men, given that measurement of prostate volume requires ultrasound or MRI.
In a more recent head to head comparison of SelectMDx and the serum 4Kscore, Wysock et al. showed SelectMDx to be inferior for detecting GG ≥2 PCa in the overall referral population. Although limited by a sample size of 50 men who underwent prostate biopsy, SelectMDx yielded an AUC of 0.67 (95% CI, 0.52–0.83) compared to the 4Kscore with an AUC of 0.83 (95% CI, 0.71–0.95) (51). The authors also report a discordance between the two biomarkers in guiding decision to biopsy, further illustrating the need for larger, prospective comparative studies to optimize clinical application.
ExoDx Prostate Intelliscore (EPI)
The Exosome Diagnostics EPI is a three-gene urinary assay that incorporates PCA3, ETS transcription factor ERG (ERG), and SAM pointed domain-containing ETS transcription factor (SPDEF) mRNA into a single numerical value (from 0 to 100) for detecting GG ≥2 disease.
In 2009, Nilsson et al. described use of urinary exosomes to detect PCa biomarkers in 11 men with PCa. The authors successfully detected PCA3 and TMPRSS2:ERG, two biomarkers with known specificity for PCa (56). Utilizing this principle, Donovan et al. developed a novel two-gene signature termed the EXO106 score (urinary PCA3 and ERG mRNA). This approach was novel in that urinary biomarker detection did not require pre-collection DRE. In 195 biopsy-naïve men with PSA 2–10 ng/mL, an EXO106 cutoff of 10 provided high sensitivity (95%), specificity (50%), NPV (98%), and PPV (35%) for GG ≥2 PCa (AUC 0.76). The EXO106 model appeared to improve upon the standard of care (SOC) model of PSA, age, race, and family history of PCa (AUC 0.67), and combining EXO106 with SOC increased the AUC to 0.80 (57).
A key EPI validation study was conducted by McKiernan and colleagues in 2016 (58), and a 2018 utility study (59) included validation data relevant to this review (Table 6).
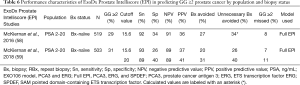
Full table
Both studies included independent cohorts of men with PSA levels ranging from 2–20 ng/mL presenting for initial biopsy (N=519 and N=503, respectively). In both cohorts, an EPI cutoff of 15.6 yielded similar sensitivity (92–93%), specificity (26–34%), NPV (89–91%), PPV (36–37%). Clinically, these data translated to avoidance of 20–27% of biopsies, while delaying the diagnosis of 7–8% of GG ≥2 cancers (58,59). These data support reproducibility and the potential use of EPI within this patient population.
MyProstateScore (MPS)
Previously named Mi-Prostate Score (MiPS), the clinically-available MyProstateScore (MPS) combines urinary PCA3 and T2:ERG with serum PSA in a validated model to predict GG ≥2 PCa (60). The resulting output is a continuous score from 0 (very unlikely to detect GG ≥2 PCa) to 100 (very likely to detect GG ≥2 PCa).
Individually, urinary PCA3 has been extensively studied and is FDA-approved for use in the repeat biopsy setting (55,61,62). Discovered in 2005, the T2:ERG gene fusion has been well-studied in tissue, where it has >99% specificity for cancer (63-65). Urinary detection of T2:ERG was achieved using a similar approach to PCA3, and urinary T2:ERG has been associated with clinically-significant PCa in subsequent studies (66,67). Combining urinary PCA3 and T2:ERG in an ‘either-or’ approach, Sanda et al. reported 93% sensitivity, 33% specificity, 93% NPV, and 33% PPV for GG ≥2 disease. Application of the combined testing approach would have avoided 42% of unnecessary biopsies at the expense of missing 7% of GG ≥2 cancers (68).
In a large multicenter study, Tomlins and colleagues derived the multivariable MPS model to optimally combine PSA, PCA3, and T2:ERG for detecting GG ≥2 cancer. MPS was subsequently applied to an external validation cohort of 1,244 men with median PSA of 4.7 (IQR, 3.3–6.5), of which 20% had a history of a prior negative biopsy. On validation, MPS provided superior predictive accuracy (AUC 0.77) for GG ≥2 PCa relative to PSA (AUC 0.65) and the PCPT-RC (AUC 0.71). Across various threshold values, use of MPS would have resulted in substantial reduction in prostate biopsy, while missing only 1.0–2.3% of GG ≥2 cancers (60).
More recently, additional multi-institutional efforts have sought to establish a pragmatic approach to MPS testing. In the biopsy-naïve setting, the MPS threshold of 10 was applied to two external validation cohorts—one in the community setting and one in the academic setting. In the combined validation data (n=1,525), the MPS threshold of 10 provided 98% negative predictive value and 97% sensitivity for GG ≥2 cancer. These findings were confirmed in 1,242 patients meeting testing criteria consistent with the National Comprehensive Cancer Network (i.e., PSA 3–10 ng/mL or PSA <3 and suspicious DRE) and in clinically-pertinent subgroups (i.e., African-American men and men with suspicious DRE). Applied as a reflex clinical test, MPS would have prevented 33% of unnecessary biopsies while failing to detect only 3.0% of GG ≥2 cancers (69). These data are listed in Table 7.

Full table
Commentary
Several serum and urine biomarkers have been proposed to improve detection of clinically-significant PCa and better inform clinical decision-making. These commercially-available tests have been validated to varying degrees in pertinent testing populations, and each appears to add diagnostic information beyond baseline clinical data.
Although promising, optimal application of these biomarkers remains to be determined. Continued work to refine the role of molecular biomarkers in prostate cancer early detection is ongoing, including efforts combining these markers with pre-biopsy prostate MRI. MRI has been used for risk stratification in the biopsy referral population and has been associated with improved detection of clinically significant PCa and reduced overdiagnosis of GG1 disease (70,71). However, the tradeoff in terms of significant cancers missed is concerning. Available data from largely expert centers cite a pooled NPV of 91% for clinically significant cancer in biopsy-naïve men but acknowledges significant heterogeneity across centers, with NPV as low as 63% (72). Variability in MRI accuracy exists within and across institutions. There is evidence supporting a wide variation in PIRADS score and cancer yield for individual readers, with clinically significant cancer detection rates ranging from 40% to 80% for lesions read as PIRADS 5 across radiologists at a single academic center (73). Furthermore, wide ranges in prostate MRI accuracy have been reported across institutions. For example, Westphalen et al. reported a PPV ranging from 35% to 49% in data collected from centers participating in the Society of Abdominal Radiology Prostate Cancer disease-focused panel, which comprises experts dedicated to prostate cancer imaging (74). These data raise concerns in using MRI as an initial secondary test for PCa evaluation, and support consideration of objective testing platforms not dependent on reader expertise.
Practically, objective tests obtained in the course of standard urologic care provide notable advantages. We have reviewed several candidate markers for the detection of clinically significant prostate cancer. Acknowledging the limitations of cross-study comparisons, these markers appear to provide diagnostic accuracy on par with that of MRI obtained at expert centers. On the contrary, MRI provides a unique ability to target high-risk lesions and is currently supported by a more robust body of prospective data than most of the biomarkers described herein (70,71,75,76). These assays stand to benefit from additional prospective studies to identify clear approaches for clinical use in better-defined testing populations (i.e., biopsy-naïve men, those with a history of negative biopsy, and African American men). Certainly, as a combined approach, initial use of biomarkers to rule out one-quarter to one-third of unnecessary biopsies, followed by MRI to improve the diagnostic yield of invasive biopsy, is highly appealing. A number of clinical trials assessing the value of MRI and biomarkers are currently underway (77,78).
While the use of imaging and biomarker-based tools appear to provide clinical benefit relative to PSA alone (71,79), their impact on the cost of care is not fully characterized. In a 2018 cost-effectiveness analysis, Sathianathen et al. found that the use of biomarkers following elevated PSA reduced unnecessary biopsies by 24% to 34% and improved quality-adjusted survival relative to the standard of care (80). Currently, the cost and availability of these tools vary across practices and reimbursement policies. As practice patterns and policies are better established, further analyses will better define the cost-effectiveness of these diagnostic modalities in addition to their clinical accuracy. Ultimately, the ability to use these tests will likely depend on practical considerations such as reimbursement, which is similarly tied to clinical evidence. Thus, the ability to optimally apply available tools will depend on the production of quality data to demonstrate a measurable impact on meaningful clinical outcomes.
Conclusions
The goal of secondary testing is to reduce the harms associated with PSA-based screening while preserving its potential life-prolonging benefit. Multiple serum and urinary biomarkers have been validated for use in avoiding unnecessary biopsies in a proportion of men, while failing to detect a limited number of GG ≥2 cancers. Emerging data focused on clear applications of these markers in specific clinical settings and populations will better define their use and support more widespread adoption. As our shared goal remains to minimize harm due to prostate cancer and improve the patient experience, thoughtfully-designed and well-executed clinical research is essential to reaching this goal.
Acknowledgments
We would like to thank Dr. Bruce J. Trock (Professor, Johns Hopkins Brady Urological Institute, Baltimore, MD) for his expert review of statistical content.
Funding: JJT is supported in-part by a University of Michigan Precision Health Research Scholar Award, the SPORE Career Enhancement Program (CA186786), and a Prostate Cancer Foundation Young Investigator Award. TMM is supported by the A. Alfred Taubman Medical Research Institute.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Badrinath R. Konety, Daniel W. Lin) for the series “Current and Future Topics on Prostate Cancer” published in Translational Andrology and Urology. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1151). The series “Current and Future Topics on Prostate Cancer” was commissioned by the editorial office without any funding or sponsorship. JJT is a co-founder with equity interest and has received stipend/honoraria from LynxDx, Inc. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144:1941-53. [Crossref] [PubMed]
- Fitzmaurice C, Akinyemiju TF, Al Lami FH, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016 a systematic analysis for the global burden of disease study global burden o. JAMA Oncol 2018;4:1553-68. [Crossref] [PubMed]
- Rawla P. Epidemiology of Prostate Cancer. World J Oncol 2019;10:63-89. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet 2016;387:70-82. [Crossref] [PubMed]
- Hugosson J, Roobol MJ, Mansson M, et al. A 16-yr Follow-up of the European Randomized study of Screening for Prostate Cancer. Eur Urol 2019;76:43-51. [Crossref] [PubMed]
- Schröder FH, Hugosson J, Roobol MJ, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med 2012;366:981-90. [Crossref] [PubMed]
- Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate cancer mortality: Results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet 2014;384:2027-35. [Crossref] [PubMed]
- Bozeman CB, Carver BS, Eastham JA, et al. Treatment of chronic prostatitis lowers serum prostate specific antigen. J Urol 2002;167:1723-6. [Crossref] [PubMed]
- Punglia RS, D’Amico AV, Catalona WJ, et al. Impact of age, benign prostatic hyperplasia, and cancer on prostate-specific antigen level. Cancer 2006;106:1507-13. [Crossref] [PubMed]
- Fenton JJ, Weyrich MS, Durbin S, et al. Prostate-Specific Antigen-Based Screening for Prostate Cancer: Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2018;319:1914-31. [Crossref] [PubMed]
- Loeb S, Bjurlin MA, Nicholson J, et al. Overdiagnosis and overtreatment of prostate cancer. Eur Urol 2014;65:1046-55. [Crossref] [PubMed]
- Loeb S, Vellekoop A, Ahmed HU, et al. Systematic review of complications of prostate biopsy. Eur Urol 2013;64:876-92. [Crossref] [PubMed]
- Barry MJ. Clinical practice. Prostate-specific-antigen testing for early diagnosis of prostate cancer. N Engl J Med 2001;344:1373-7. [Crossref] [PubMed]
- Welch HG, Fisher ES, Gottlieb DJ, et al. Detection of prostate cancer via biopsy in the Medicare-SEER population during the PSA era. J Natl Cancer Inst 2007;99:1395-400. [Crossref] [PubMed]
- Lamy PJ, Allory Y, Gauchez AS, et al. Prognostic Biomarkers Used for Localised Prostate Cancer Management: A Systematic Review. Eur Urol Focus 2018;4:790-803. [Crossref] [PubMed]
- National Comprehensive Cancer Network. Prostate Cancer Early Detection (Version 2.2020) [Internet]. Available online: https://www.nccn.org/professionals/physician_gls/PDF/prostate_detection.pdf
- Trevethan R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front Public Health 2017;5:307. [Crossref] [PubMed]
- Tosoian JJ, Ross AE, Sokoll LJ, et al. Urinary Biomarkers for Prostate Cancer. Urol Clin North Am 2016;43:17-38. [Crossref] [PubMed]
- Florkowski CM. Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: communicating the performance of diagnostic tests. Clin Biochem Rev 2008;29:S83-7. [PubMed]
- Janes H, Pepe MS. Adjusting for covariates in studies of diagnostic, screening, or prognostic markers: An old concept in a new setting. Am J Epidemiol 2008;168:89-97. [Crossref] [PubMed]
- Pepe MS, Fan J, Seymour CW, et al. Biases introduced by choosing controls to match risk factors of cases in biomarker research. Clin Chem 2012;58:1242-51. [Crossref] [PubMed]
- Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010;21:128-38. [Crossref] [PubMed]
- Van Calster B, Wynants L, Verbeek JFM, et al. Reporting and Interpreting Decision Curve Analysis: A Guide for Investigators. Eur Urol 2018;74:796-804. [Crossref] [PubMed]
- Catalona WJ, Partin AW, Slawin KM, et al. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: a prospective multicenter clinical trial. JAMA 1998;279:1542-7. [Crossref] [PubMed]
- Catalona WJ, Partin AW, Sanda MG, et al. A multicenter study of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ml prostate specific antigen range. J Urol 2011;185:1650-5. [Crossref] [PubMed]
- Fossati N, Lazzeri M, Haese A, et al. Clinical performance of serum isoform [-2]proPSA (p2PSA), and its derivatives %p2PSA and the Prostate Health Index, in men aged <60 years: results from a multicentric European study. BJU Int 2015;115:913-20. [Crossref] [PubMed]
- Lazzeri M, Briganti A, Scattoni V, et al. Serum index test %[-2]proPSA and Prostate Health Index are more accurate than prostate specific antigen and %fPSA in predicting a positive repeat prostate biopsy. J Urol 2012;188:1137-43. [Crossref] [PubMed]
- Foley RW, Gorman L, Sharifi N, et al. Improving multivariable prostate cancer risk assessment using the Prostate Health Index. BJU Int 2016;117:409-17. [Crossref] [PubMed]
- Stephan C, Vincendeau S, Houlgatte A, et al. Multicenter evaluation of [-2]proprostate-specific antigen and the prostate health index for detecting prostate cancer. Clin Chem 2013;59:306-14. [Crossref] [PubMed]
- Guazzoni G, Nava L, Lazzeri M, et al. Prostate-specific antigen (PSA) isoform p2PSA significantly improves the prediction of prostate cancer at initial extended prostate biopsies in patients with total PSA between 2.0 and 10 ng/ml: results of a prospective study in a clinical setting. Eur Urol 2011;60:214-22. [Crossref] [PubMed]
- Ferro M, Bruzzese D, Perdona S, et al. Prostate Health Index (Phi) and Prostate Cancer Antigen 3 (PCA3) significantly improve prostate cancer detection at initial biopsy in a total PSA range of 2-10 ng/ml. PLoS One 2013;8:e67687 [Crossref] [PubMed]
- Lazzeri M, Haese A, de la Taille A, et al. Serum isoform [-2]proPSA derivatives significantly improve prediction of prostate cancer at initial biopsy in a total PSA range of 2-10 ng/ml: a multicentric European study. Eur Urol 2013;63:986-94. [Crossref] [PubMed]
- Loeb S, Sokoll LJ, Broyles DL, et al. Prospective multicenter evaluation of the Beckman Coulter Prostate Health Index using WHO calibration. J Urol 2013;189:1702-6. [Crossref] [PubMed]
- Boegemann M, Stephan C, Cammann H, et al. The percentage of prostate-specific antigen (PSA) isoform [-2]proPSA and the Prostate Health Index improve the diagnostic accuracy for clinically relevant prostate cancer at initial and repeat biopsy compared with total PSA and percentage free PSA in men. BJU Int 2016;117:72-9. [Crossref] [PubMed]
- de La Calle C, Patil D, Wei JT, et al. Multicenter evaluation of the prostate health index to detect aggressive prostate cancer in biopsy Naïve men. J Urol 2015;194:65-72. [Crossref] [PubMed]
- Nordström T, Vickers A, Assel M, et al. Comparison Between the Four-kallikrein Panel and Prostate Health Index for Predicting Prostate Cancer. Eur Urol 2015;68:139-46. [Crossref] [PubMed]
- Seisen T, Roupret M, Brault D, et al. Accuracy of the prostate health index versus the urinary prostate cancer antigen 3 score to predict overall and significant prostate cancer at initial biopsy. Prostate 2015;75:103-11. [Crossref] [PubMed]
- Chiu PK, Ng CF, Semjonow A, et al. A Multicentre Evaluation of the Role of the Prostate Health Index (PHI) in Regions with Differing Prevalence of Prostate Cancer: Adjustment of PHI Reference Ranges is Needed for European and Asian Settings. Eur Urol 2019;75:558-61. [Crossref] [PubMed]
- Loeb S, Sanda MG, Broyles DL, et al. The prostate health index selectively identifies clinically significant prostate cancer. J Urol 2015;193:1163-9. [Crossref] [PubMed]
- FDA Premarket Approval Application (PMA) Number: P090026. [Internet]. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf9/P090026b.pdf
- EAU Guidelines. Edn. presented at the EAU Annual Congress Copenhagen. In 2018.
- Vickers AJ, Cronin AM, Aus G, et al. A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: data from the European Randomized Study of Prostate Cancer Screening in Goteborg, Sweden. BMC Med 2008;6:19. [Crossref] [PubMed]
- Vickers A, Cronin A, Roobol M, et al. Reducing unnecessary biopsy during prostate cancer screening using a four-kallikrein panel: an independent replication. J Clin Oncol 2010;28:2493-8. [Crossref] [PubMed]
- Vickers AJ, Cronin AM, Aus G, et al. Impact of recent screening on predicting the outcome of prostate cancer biopsy in men with elevated prostate-specific antigen: data from the European Randomized Study of Prostate Cancer Screening in Gothenburg, Sweden. Cancer 2010;116:2612-20. [Crossref] [PubMed]
- Vickers AJ, Cronin AM, Roobol MJ, et al. A four-kallikrein panel predicts prostate cancer in men with recent screening: data from the European Randomized Study of Screening for Prostate Cancer, Rotterdam. Clin Cancer Res 2010;16:3232-9. [Crossref] [PubMed]
- Parekh DJ, Punnen S, Sjoberg DD, et al. A Multi-institutional Prospective Trial in the USA Confirms that the 4Kscore Accurately Identifies Men with High-grade Prostate Cancer. Eur Urol 2015;68:464-70. [Crossref] [PubMed]
- Braun K, Sjoberg DD, Vickers AJ, et al. A Four-kallikrein Panel Predicts High-grade Cancer on Biopsy: Independent Validation in a Community Cohort. Eur Urol 2016;69:505-11. [Crossref] [PubMed]
- Punnen S, Freedland SJ, Polascik TJ, et al. A Multi-Institutional Prospective Trial Confirms Noninvasive Blood Test Maintains Predictive Value in African American Men. J Urol 2018;199:1459-63. [Crossref] [PubMed]
- Haese A, Trooskens G, Steyaert S, et al. Multicenter Optimization and Validation of a 2-Gene mRNA Urine Test for Detection of Clinically Significant Prostate Cancer before Initial Prostate Biopsy. J Urol 2019;202:256-63. [Crossref] [PubMed]
- Wysock JS, Becher E, Persily J, et al. Concordance and Performance of 4Kscore and SelectMDx for Informing Decision to Perform Prostate Biopsy and Detection of Prostate Cancer. Urology 2020;141:119-24. [Crossref] [PubMed]
- Zappala SM, Dong Y, Linder V, et al. The 4Kscore blood test accurately identifies men with aggressive prostate cancer prior to prostate biopsy with or without DRE information. Int J Clin Pract 2017;71. [Crossref] [PubMed]
- Bryant RJ, Sjoberg DD, Vickers AJ, et al. Predicting high-grade cancer at ten-core prostate biopsy using four kallikrein markers measured in blood in the ProtecT study. J Natl Cancer Inst 2015;107:djv095 [Crossref] [PubMed]
- Leyten GH, Hessels D, Smit FP, et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin Cancer Res 2015;21:3061-70. [Crossref] [PubMed]
- Van Neste L, Hendriks RJ, Dijkstra S, et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur Urol 2016;70:740-8. [Crossref] [PubMed]
- Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 2009;100:1603-7. [Crossref] [PubMed]
- Donovan MJ, Noerholm M, Bentink S, et al. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis 2015;18:370-5. [Crossref] [PubMed]
- McKiernan J, Donovan MJ, O’Neill V, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol 2016;2:882-9. [Crossref] [PubMed]
- McKiernan J, Donovan MJ, Margolis E, et al. A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer in Patients with Prostate-specific Antigen 2-10ng/ml at Initial Biopsy. Eur Urol 2018;74:731-8. [Crossref] [PubMed]
- Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur Urol 2016;70:45-53. [Crossref] [PubMed]
- Wei JT, Feng Z, Partin AW, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol 2014;32:4066-72. [Crossref] [PubMed]
- Merdan S, Tomlins SA, Barnett CL, et al. Assessment of long-term outcomes associated with urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion at repeat biopsy. Cancer 2015;121:4071-9. [Crossref] [PubMed]
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644-8. [Crossref] [PubMed]
- Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 2008;10:177-88. [Crossref] [PubMed]
- Tomlins SA, Aubin SM, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med 2011;3:94ra72 [Crossref] [PubMed]
- Salami SS, Schmidt F, Laxman B, et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol 2013;31:566-71. [Crossref] [PubMed]
- Leyten GH, Hessels D, Jannink SA, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol 2014;65:534-42. [Crossref] [PubMed]
- Sanda MG, Feng Z, Howard DH, et al. Association between combined TMPRSS2:ERG and PCA3 RNA urinary testing and detection of aggressive prostate cancer. JAMA Oncol 2017;3:1085-93. [Crossref] [PubMed]
- Tosoian JJ, Trock BJ, Morgan TM, et al. Use of the MyProstateScore Test to Rule Out Clinically Significant Cancer: Validation of a Straightforward Clinical Testing Approach. J Urol 2021;205:732-9. [Crossref] [PubMed]
- Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015;313:390-7. [Crossref] [PubMed]
- Ahdoot M, Wilbur AR, Reese SE, et al. MRI-Targeted, Systematic, and Combined Biopsy for Prostate Cancer Diagnosis. N Engl J Med 2020;382:917-28. [Crossref] [PubMed]
- Sathianathen NJ, Omer A, Harriss E, et al. Negative Predictive Value of Multiparametric Magnetic Resonance Imaging in the Detection of Clinically Significant Prostate Cancer in the Prostate Imaging Reporting and Data System Era: A Systematic Review and Meta-analysis. Eur Urol 2020;78:402-14. [Crossref] [PubMed]
- Sonn GA, Fan RE, Ghanouni P, et al. Prostate Magnetic Resonance Imaging Interpretation Varies Substantially Across Radiologists. Eur Urol Focus 2019;5:592-9. [Crossref] [PubMed]
- Westphalen AC, McCulloch CE, Anaokar JM, et al. Variability of the Positive Predictive Value of PI-RADS for Prostate MRI across 26 Centers: Experience of the Society of Abdominal Radiology Prostate Cancer Disease-focused Panel. Radiology 2020;296:76-84. [Crossref] [PubMed]
- Ahmed HU, El-Shater Bosaily A, Brown LC, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 2017;389:815-22. [Crossref] [PubMed]
- Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med 2018;378:1767-77. [Crossref] [PubMed]
- Predicting Prostate Biopsy Results With Biomarkers and mpMRI. (ProBioM) [Internet]. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US).; Available online: https://clinicaltrials.gov/show/NCT03730324
- EDRN Prostate MRI Biomarker Study (P-MRI) [Internet]. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US); Available online: https://clinicaltrials.gov/show/NCT03784924
- Gulati R, Morgan TM, A’Mar T, et al. Overdiagnosis and Lives Saved by Reflex Testing Men with Intermediate Prostate-Specific Antigen Levels. J Natl Cancer Inst 2020;112:384-90. [Crossref] [PubMed]
- Sathianathen NJ, Kuntz KM, Alarid-Escudero F, et al. Incorporating Biomarkers into the Primary Prostate Biopsy Setting: A Cost-Effectiveness Analysis. J Urol 2018;200:1215-20. [Crossref] [PubMed]


