
Construction and analysis of a ceRNA network and patterns of immune infiltration in bladder cancer
Introduction
Bladder cancer (BC) is the ninth most common malignant tumor worldwide (1), accounting for an estimate of 549,000 new BC cases and 200,000 BC-related deaths in 2018 (2). BC is classified into non-muscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC), and transitional cell carcinoma is the most common pathological type (3,4). NMIBC accounts for 75% of BC cases, and 50% of NMIBC cases progress to MIBC (5). Transurethral resection of bladder tumor (TURBt) is considered the main treatment for NMIBC, and radical cystectomy is the strategy for MIBC treatment (6). However, current treatment strategies for BC have not significantly improved the 5-year survival rate over the past decade (7). Moreover, BC has become one of the most expensive to treat solid tumors due to high recurrence rate (8). Therefore, it is essential to identify biomarkers that can accurately evaluate the diagnosis, treatment, and prognosis of BC.
Noncoding RNAs (ncRNAs), including long noncoding RNAs (lncRNAs) and microRNAs (miRNAs), have been recently recognized as the key regulatory factors in tumors (9). Overexpression of lncRNA DLX6 antisense RNA 1 (DLX6-AS1) and insulin-like growth factor binding protein 4-1 (IGFBP4-1) may lead to a poor prognosis in BC patients (10,11). Upregulation of miRNA-133b and miR-375-3p inhibits the proliferation and metastasis of BC cells (12,13). The occurrence of tumors is closely related to interactions between lncRNAs, mRNAs, and miRNAs (14). LncRNAs and mRNAs can competitively bind miRNAs by miRNA response elements (MREs) to generate a competitive endogenous RNA (ceRNA) network to regulate the expression of various RNAs and proteins (15,16). This ceRNA network can be used to investigate the functions of lncRNAs and to identify biomarkers related to the diagnosis and prognosis of BC, which is essential for assisting the clinicians with early diagnosis, risk assessment, and appropriate treatment decision-making for patients with BC (17,18).
Immunotherapy has become a promising antitumor strategy that recognizes tumors as foreign antigens and inhibits the proliferation and metastasis of the tumor cells by inducing active or passive immune responses (19,20). In BC, Bacillus Calmette-Guerin vaccine (BCG) suppresses tumor cells by activating immune cell infiltration and is the gold standard for therapy of high-risk NMIBC (21,22). However, approximately 40% of BC patients do not respond to BCG, and 15% of BC patients experience progression to MIBC after the treatment (23,24). Recent studies have shown that immune-related genes were significantly associated with the prognosis of cancer. High expression of ENDRA was associated with lower survival rate in patients with advanced BC (25). Overexpression of RAC3 promoted the migration of cancer cells and was associated with a decrease in the recurrence-free survival in ER3-positive breast cancer (26). The exploration of immune checkpoint inhibitors (ICIs) facilitated great progress in immunotherapy. In phase II clinical trials, the neoadjuvant use of ICIs resulted in pathological complete responses in patients with MIBC (27).
A number of recent studies demonstrated important roles of the ceRNA networks and immune cell infiltration in evaluation of prognosis in patients with malignant tumors (28,29). Some studies suggested that a combination of a ceRNA network and immune cell infiltration has potential value in cancer research (30,31). Therefore, we constructed a ceRNA network for BC using the data downloaded from The Cancer Genome Atlas (TCGA) database. Two survival analyses were performed, including analysis based on selected genes included in the ceRNA network and analysis based on the data on immune cell infiltration estimated by the Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) algorithm. These two survival analyses were used to identify the key factors and immune cells that can accurately predict patients with BC, and these factors were combined for subsequent analysis, which is very important for early diagnosis and treatment of BC. Finally, the relationships between these two survival analyses were analyzed (Figure 1). The article is presented in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1250).
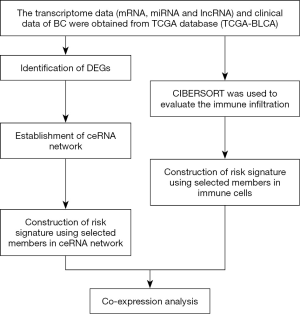
Methods
Identification of differentially expressed genes (DEGs)
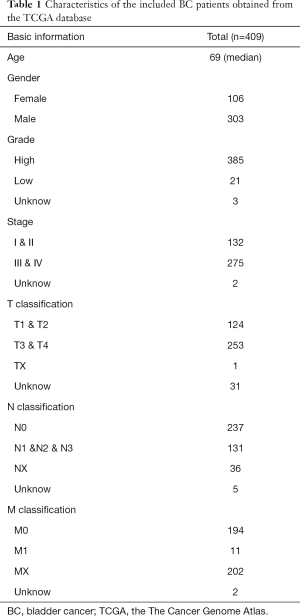
The data for 411 BC tissues and 19 normal tissues were downloaded from the TCGA dataset (The Cancer Genome Atlas) (https://portal.gdc.cancer.gov/) and used for mRNA and lncRNA analysis. The data for 418 BC tissues and 19 normal tissues were downloaded from the TCGA dataset and used for miRNA analysis. The clinical data of 409 patients with BC were obtained from TCGA (including age, gender, grade, stage, and TNM classification) (Table 1). The DESeq2 method was used to identify DEGs, including lncRNAs, miRNAs, and mRNAs. DESeq2 can improve the stability and interpretability of estimation fold change (FC) and dispersion by shrinkage estimators (32,33). The data corresponding to duplicate and abnormal samples (not primary tumors and abnormal samples) were deleted, and the remaining data were normalized by the trimmed mean of M-values (TMM) method implemented in edgeR and further transformed by the voom method provided in limma. TMM normalization is a simple and effective method for estimation of relative RNA levels using RNA sequencing (RNA-seq) data. Moreover, voom is faster and more convenient than existing methods of RNA-seq analysis and converts RNA-seq data into a form that can be analyzed using tools similar to those used for microarray analysis. The cutoff criteria were defined as |log2 FC| >1.0 and false discovery rate (FDR) adjusted P<0.05. Ethical approval was not required because the data used in the present study were obtained from public databases. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Full table
Construction of a ceRNA network
StarBase was introduced to predict the lncRNA–miRNA and miRNA–mRNA interactions (34). StarBase (http://starbase.sysu.edu.cn/) is an open-source database that can display extensive and complex RNA-RNA (miRNA-lncRNA, miRNA-pseudogene, miRNA-circRNA, and miRNA-mRNA) and protein-RNA interaction networks by analyzing millions binding sites of Argonaute (Ago) and other RNA-binding protein (RBP) identified by crosslinking immunoprecipitation and high-throughput sequencing (CLIP-seq) (35). Interactions with P values <0.05 for the hypergeometric and correlation tests were considered significant. We used Cytoscape software version 3.8.0 to display the ceRNA network.
Survival analysis based on the components of the ceRNA network
All components of the ceRNA network were incorporated into a univariate Cox regression model to identify the genes related to survival (P<0.05). The survival analysis of the components of the ceRNA network was performed by least absolute shrinkage and selection operator (LASSO) Cox regression to avoid overfitting (36). The median value of the risk score calculated by LASSO was used as the cutoff value to divide patients into the high-risk and the low-risk groups. Kaplan-Meier survival curves were compared using the log-rank test and a nomogram to assess the survival outcomes [overall survival (OS)]. A receiver operating characteristic (ROC) curve and a calibration curve were constructed to evaluate the accuracy of the risk score.
Immune cell infiltration
We used CIBERSORT to evaluate the immune infiltration data for each BC sample downloaded from TCGA. CIBERSORT is a deconvolution algorithm that can predict the abundance of 22 immune cell types based on the gene expression data (37,38).
Survival analysis based on selected immune cells
All immune cells were incorporated into a univariate Cox regression model to identify immune cells related to survival (P<0.05). The survival analysis based on immune cells was performed by LASSO Cox regression to avoid overfitting. The patients were divided into the high-risk and low-risk groups based on the median value of the risk score. Kaplan-Meier survival curves were compared using the log-rank test and a nomogram to assess the survival outcomes (OS). A ROC curve and a calibration curve were constructed to evaluate the accuracy of the risk score. Finally, Pearson correlation analysis was used to assess the correlations between various immune cells and between the components of the immune cell signature and the components of the ceRNA network signature.
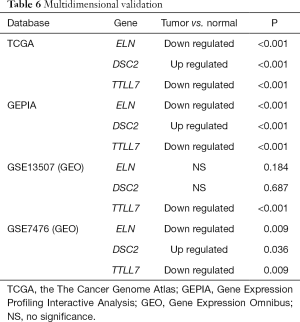
Multidimensional validation
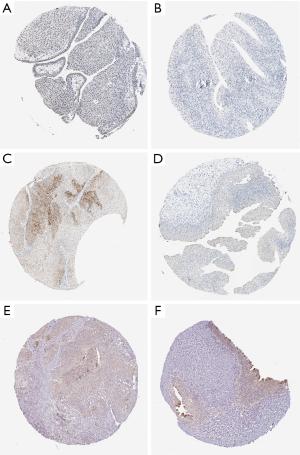
Multidimensional verification was performed using a series of external databases, including Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/), the Gene Expression Profiling Interactive Analysis (GEPIA), and TargetScan database (http://www.targetscan.org/), to reduce possible errors. We also downloaded the relevant immunohistochemical images from the public database Human Protein Atlas database (HPA) (https://www.proteinatlas.org/) to detect the gene expression in tumor tissues and normal tissues, but no specific staining method was provided in the database.
Statistical analysis
All statistical analyses were performed using R 3.53 software. A ceRNA network was constructed using the GDCRNATools package of R (39). The log-rank test and univariate Cox regression analyses were performed using the survival package of R. The survival time and status of the patients were assumed to be dependent variables, and the expression of the components of the ceRNA network and immune cell infiltration were assumed to be independent variables in Cox regression used for two survival analyses. LASSO Cox regression was performed using the glmnet package of R. ROC curves were generated and plotted using the survivalROC package of R. Nomograms and calibration curves were generated using the rms package of R. The differences in the gene expression levels were evaluated by the Mann-Whitney U test.
Results
DEGs
We identified 2,581 differentially expressed mRNAs (1,261 upregulated and 1,320 downregulated mRNAs), 223 differentially expressed miRNAs (152 upregulated and 71 downregulated miRNAs), and 216 differentially expressed lncRNAs (39 upregulated and 177 downregulated lncRNAs) in BC versus normal tissues (Figure 2A,B,C).

Construction of a ceRNA network and survival analysis
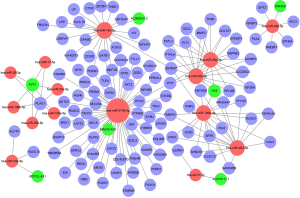
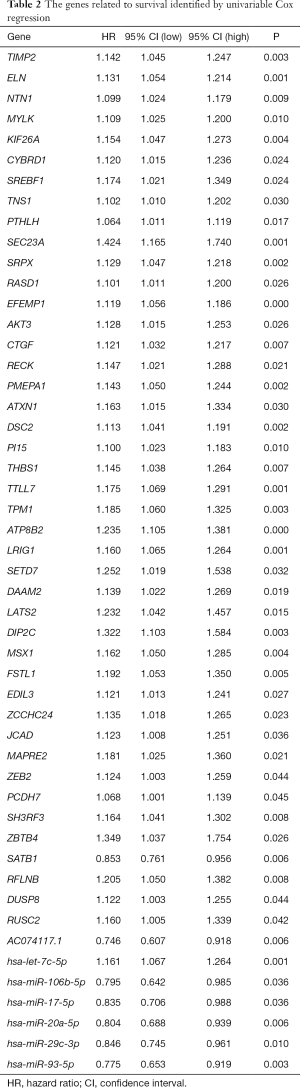
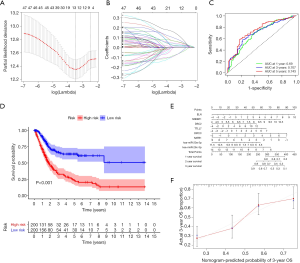
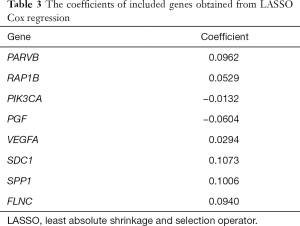
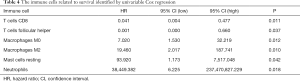
We constructed a ceRNA network composed of 110 mRNAs, 15 miRNAs, and 7 lncRNAs (Figure 3). Then, all components of the ceRNA network were incorporated into a univariate Cox regression model to identify components related to survival (P<0.05) (Table 2). The survival analysis by LASSO Cox regression used the selected components (Figure 4A,B; Table 3). The median value of the risk score (1.072) calculated by LASSO was used as the cutoff value to divide BC patients into the high-risk and low-risk groups. ROC curve showed that OS of BC patients was perfectly predicted by the risk score [area under the curve (AUC) for 1-year survival =0.690; AUC for 3-year survival =0.707; and AUC for 5-year survival =0.743] (Figure 4C). The prognosis of BC patients in the high-risk group was worse than that in the low-risk group (P<0.001) (Figure 4D). A nomogram predicted the probability of 1-, 3-, and 5-year OS (Figure 4E). The calibration curve indicated that prediction using the nomogram was accurate (Figure 4F).
Full table
Full table
Immune cell infiltration
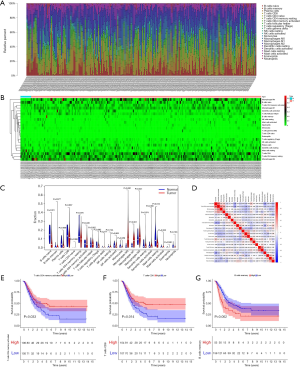
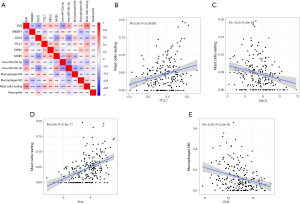
CIBERSORT was used to evaluate the immune infiltration data for each BC sample downloaded from TCGA, and bar plots and heatmaps were used to visualize the data (Figure 5A,B). The proportions of naive B cells (P=0.001), M0 macrophages (P<0.001), M1 macrophages (P<0.001), and resting mast cells (P=0.012) were different in the tumor and normal tissues (Figure 5C). Analysis of correlations between DEG expression and immune cell infiltration in BC and normal tissues (Figure 5D) indicated that high levels of CD4 memory-activated T cells (P=0.033) and CD8 T cells (P=0.014) were associated with favorable OS, and high levels of memory B cells were associated with unfavorable survival (P=0.002) (Figure 5E,F,G). Additionally, we evaluated correlations between clinical and pathological factors and immune cells (Figure 6).
Survival analysis based on immune cells
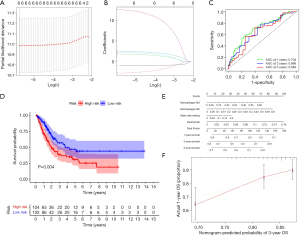
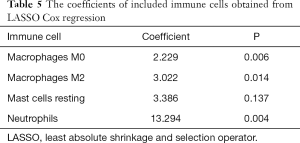
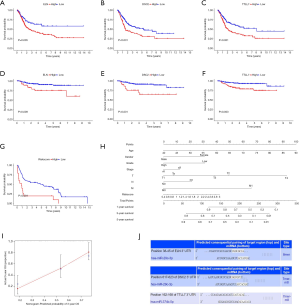
All immune cells were incorporated into a univariate Cox regression model to identify immune cells associated with survival (P<0.05) (Table 4). LASSO Cox regression was used for survival analysis to avoid overfitting (Figure 7A,B) (Table 5). The median value of the risk score (0.805) calculated by LASSO was used as the cutoff value to divide patients into the high-risk and low-risk groups. ROC curves indicated that OS of BC patients was perfectly predicted by the risk score (AUC for 1-year survival =0.704; AUC for 3-year survival =0.666; and AUC for 5-year survival =0.648) (Figure 7C). The prognosis of BC patients in the high-risk group was worse than that in the low-risk group (P=0.004) (Figure 7D). A nomogram predicted the survival probability of 1-, 3-, and 5-year OS (Figure 7E). The calibration curve confirmed that prediction using the nomogram was accurate (Figure 7F). Finally, correlations between the components of the ceRNA signature and immune cell signature were analyzed (Figure 8A). Tubulin tyrosine ligase like 7 (TTLL7) and resting mast cells (R=0.24, P<0.001), desmocollin 2 (DSC2) and resting mast cells (R=−0.23, P<0.001), elastin (ELN) and resting mast cells (R=0.44, P<0.001), and hsa-miR-29c-3p and M0 macrophages (R=−0.29, P<0.001) had significant correlations (Figure 8B,C,D,E).
Full table
Full table
Multidimensional validation
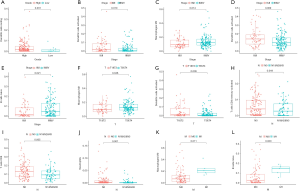
Multidimensional verification included a series of external databases (GEO, GEPIA, HPA, and TargetScan database) to verify the characteristics of significant factors in the coexpression test and survival analysis based on immune cells. Four GEO cohorts were used, including GSE13507, GSE7476, GSE32894, and GSE31684. The results obtained using the TCGA database indicated that the expression of ELN and TTLL7 was higher in normal tissues and the expression of DSC2 was higher in BC tissues. Analysis using the GEPIA database confirmed that both ELN and TTLL7 were expressed at a low level in tumor tissues, and the expression of DSC2 was upregulated. Analysis of the GSE13507 cohort indicated a lack of significant differences in the expression of ELN and DSC2, and the expression of TTLL7 was decreased in BC tissues compared with that in normal tissues. In the GSE7476 cohort, DSC2 was overexpressed and the expression of TTLL7 was decreased in BC tissues; however, the expression of ELN was not significantly different in tumor and normal tissues. The above results were summarized in Table 6. HPA database search results showed that the expression levels of ELN, DSC2, and TTLL7 were significantly increased in BC tissues (Figure 9A,B,C,D,E,F). Analysis of the data of multiple databases (TCGA and GSE31894) indicated that high levels of ELN, DSC2, and TTLL7 were significantly associated with poor prognosis of BC patients (Figure 10A,B,C,D,E,F). Finally, we used GSE31684 to validate the survival analysis based on immune cells. The prognosis of the high-risk group was worse than that of the low-risk group (P<0.001) (Figure 10G). A nomogram was able to predict the survival probability of 1-, 3-, and 5-year OS (Figure 10H). The calibration curve indicated that prediction using the nomogram was accurate (Figure 10I). Unfortunately, we were unable to verify the survival analysis based on the components of the ceRNA network due to a lack of databases that include mRNA, miRNA, lncRNA, and complete clinical data, which were present only in the TCGA database. Query of the TargetScan database identified binding sites for hsa-miR-29c-3p in ELN and DSC2, and a binding site for hsa-miR-374b-5p was identified in TTLL7 in the ceRNA network (Figure 10J).
Full table
Discussion
The number of estimated new BC cases reported in 2019 was 80,470, and 17,670 BC-related deaths were registered. BC is characterized by complex biological behavior, a high rate of metastasis, and a high recurrence rate (40,41). After diagnosis, the physical, mental, and social health-related quality of life (HRQoL) of patients with BC significantly declines (42). Increasing evidence indicated that ceRNAs and dysregulation of the immune system are involved in the progression of human tumors (43-48).
Two survival analyses were performed in the present study based on the selected components of a ceRNA network (including ELN, SREBF1, DSC2, TTLL7, DIP2C, SATB1, hsa-miR-20a-5p, and hsa-miR-29c-3p) and selected immune cells (including M0 macrophages, M2 macrophages, resting mast cells, and neutrophils). Overexpression of SREBF1 was associated with a poor prognosis and promoted metastasis of esophageal carcinoma cells, and inhibition of SREBF1 augmented the efficacy of immune checkpoint blockades (49,50). Downregulated expression of DIP2C was detected in breast cancer, especially in basal-like and HER-2 subtypes (51). Numerous studies confirmed that SATB1 is involved in the occurrence and development of various human tumors (52-55). Hsa-miR-20a-5p inhibits epithelial mesenchymal transition (EMT) and invasion of tumor cells by targeting STAT3 in endometrial cancer (56). The infiltration of M2 macrophages expressing the CD163 antigen decreases the disease-free survival (DFS) rate in high-grade oral tongue squamous cell carcinoma and increases the mortality rate of patients with prostate cancer (57,58). The interaction between neutrophils and cancer is complex. Neutrophil-mediated tumor inflammation can promote the proliferation, invasion, and angiogenesis of tumor cells; however, some studies demonstrated that compounds produced by neutrophils can suppress tumor development (59-61).
The present study demonstrated significant correlations between TTLL7 and resting mast cells, DSC2 and resting mast cells, ELN and resting mast cells, and hsa-miR-29c-3p and M0 macrophages. Another study suggested that DS2 can be used as a new immunohistochemical biomarker for urothelial carcinoma (UC) with squamous differentiation; this cancer is more advanced and has worse prognosis than those in UC without squamous differentiation (62). In breast cancer, prostate cancer, and lung cancer, increased levels of mast cell infiltration were associated with improved survival rate of patients (63). Decreased expression of hsa-miR-29c-3p was associated with advanced clinical and pathological factors and poor prognosis in laryngeal squamous cell carcinoma, and infiltration of M0 macrophages was associated with poor OS, suggesting a negative correlation between these factors in tumors (64,65). Unfortunately, other components of ceRNA signature were rarely investigated in tumors, and previous studies did not assess possible relationships of these components with immune cells.
The present study has certain limitation. First, our data were obtained from public databases, and clinical and pathological factors included in these databases were not comprehensive; the number of tumor tissue samples was different from the number of normal tissue samples. Second, specific mechanism of infiltration of various immune cell types was not studied in detail. Third, the correlations between the components of the ceRNA network signature and the immune infiltrating cell signature were not confirmed in experiments.
Thus, two survival analyses performed in the present study were based on the selected components of the ceRNA network and selected immune cell types and were used to predict the prognosis of patients with BC; the nomograms containing both of these signatures may provide assistance to clinicians to improve individual management of BC patients. Moreover, significant correlations between TTLL7 and resting mast cells, DSC2 and resting mast cells, ELN and resting mast cells, and hsa-miR-29c-3p and M0 macrophages were detected, suggesting that these correlations play significant roles in the prognosis of BC. The present study provides some useful information for prediction of the prognosis of BC patients; however, further studies are needed to clarify the relationships between ceRNAs and immune infiltrating cells and relevant molecular mechanisms.
Acknowledgments
Funding: This work was supported by the Project of Liaoning Distinguished Professor [grant no. [2012]145] for KCZ, the Shenyang Plan Project of Science and Technology [grant no. F17-230-9-08] for KCZ, China Medical University 2017 Discipline Promotion Program [grant no. 3110117040] for KCZ, China Medical University 2018 Discipline Promotion Program for KCZ, and the 2017 National Key R&D Program Key Projects of Precision Medical Research [no. 2017YFC0908000] for KCZ.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1250
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1250). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Cheng S, Jiang Z, Xiao J, et al. The prognostic value of six survival-related genes in bladder cancer. Cell Death Discov 2020;6:58. [Crossref] [PubMed]
- Wang Y, Chen L, Yu M, et al. Immune-related signature predicts the prognosis and immunotherapy benefit in bladder cancer. Cancer Med 2020;9:7729-41. [Crossref] [PubMed]
- Nabavizadeh R, Bobrek K, Master VA. Risk stratification for bladder cancer: Biomarkers of inflammation and immune activation. Urol Oncol 2020;38:706-12. [Crossref] [PubMed]
- Li F, Guo H, Wang Y, et al. Profiles of tumor-infiltrating immune cells and prognostic genes associated with the microenvironment of bladder cancer. Int Immunopharmacol 2020;85:106641 [Crossref] [PubMed]
- Chen X, Jin Y, Gong L, et al. Bioinformatics Analysis Finds Immune Gene Markers Related to the Prognosis of Bladder Cancer. Front Genet 2020;11:607. [Crossref] [PubMed]
- Lucca I, de Martino M, Klatte T, et al. Novel biomarkers to predict response and prognosis in localized bladder cancer. Urol Clin North Am 2015;42:225-33. ix. [Crossref] [PubMed]
- Zhang XQ, Sun S, Lam KF, et al. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis 2013;58:123-31. [Crossref] [PubMed]
- Wang H, Niu X, Jiang H, et al. Long non-coding RNA DLX6-AS1 facilitates bladder cancer progression through modulating miR-195-5p/VEGFA signaling pathway. Aging (Albany NY) 2020;12:16021-34. [Crossref] [PubMed]
- Li C, Cao Y, Zhang L, et al. LncRNA IGFBP4-1 promotes tumor development by activating Janus kinase-signal transducer and activator of transcription pathway in bladder urothelial carcinoma. Int J Biol Sci 2020;16:2271-82. [Crossref] [PubMed]
- Li Q, Huyan T, Cai S, et al. The role of exosomal miR-375-3p: A potential suppressor in bladder cancer via the Wnt/β-catenin pathway. FASEB J 2020;34:12177-96. [Crossref] [PubMed]
- Cai X, Qu L, Yang J, et al. Exosome-transmitted microRNA-133b inhibited bladder cancer proliferation by upregulating dual-specificity protein phosphatase 1. Cancer Med 2020;9:6009-19. [Crossref] [PubMed]
- Zhu X, Jiang L, Yang H, et al. Analyzing the lncRNA, miRNA, and mRNA-associated ceRNA networks to reveal potential prognostic biomarkers for glioblastoma multiforme. Cancer Cell Int 2020;20:393. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Long J, Bai Y, Yang X, et al. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int 2019;19:90. [Crossref] [PubMed]
- Jiang Y, Wang J, Chen J, et al. Construction and analysis of an aberrant lncRNA-miRNA-mRNA network associated with papillary thyroid cancer. Medicine (Baltimore) 2020;99:e22705 [Crossref] [PubMed]
- Wang Z, Li Y, Zhong Y, et al. Comprehensive Analysis of Aberrantly Expressed Competitive Endogenous RNA Network and Identification of Prognostic Biomarkers in Pheochromocytoma and Paraganglioma. Onco Targets Ther 2020;13:11377-95. [Crossref] [PubMed]
- Carter BW, Halpenny DF, Ginsberg MS, et al. Immunotherapy in Non-Small Cell Lung Cancer Treatment: Current Status and the Role of Imaging. J Thorac Imaging 2017;32:300-12. [Crossref] [PubMed]
- Lin P, Guo YN, Shi L, et al. Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging (Albany NY) 2019;11:480-500. [Crossref] [PubMed]
- Kresowik TP, Griffith TS. Bacillus Calmette-Guerin immunotherapy for urothelial carcinoma of the bladder. Immunotherapy 2009;1:281-8. [Crossref] [PubMed]
- Pettenati C, Ingersoll MA. Mechanisms of BCG immunotherapy and its outlook for bladder cancer. Nat Rev Urol 2018;15:615-25. [Crossref] [PubMed]
- Herr HW, Morales A. History of bacillus Calmette-Guerin and bladder cancer: an immunotherapy success story. J Urol 2008;179:53-6. [Crossref] [PubMed]
- Seidl C. Targets for Therapy of Bladder Cancer. Semin Nucl Med 2020;50:162-70. [Crossref] [PubMed]
- Laurberg JR, Jensen JB, Schepeler T, et al. High expression of GEM and EDNRA is associated with metastasis and poor outcome in patients with advanced bladder cancer. BMC Cancer 2014;14:638. [Crossref] [PubMed]
- Walker MP, Zhang M, Le TP, et al. RAC3 is a pro-migratory co-activator of ERα. Oncogene 2011;30:1984-94. [Crossref] [PubMed]
- Gust KM, Rebhan K, Resch I, et al. Immune checkpoint inhibition in muscle-invasive and locally advanced bladder cancer. Curr Opin Urol 2020;30:547-56. [Crossref] [PubMed]
- Zhu N, Hou J, Wu Y, et al. Integrated analysis of a competing endogenous RNA network reveals key lncRNAs as potential prognostic biomarkers for human bladder cancer. Medicine (Baltimore) 2018;97:e11887 [Crossref] [PubMed]
- Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960-4. [Crossref] [PubMed]
- Huang R, Wu J, Zheng Z, et al. The Construction and Analysis of ceRNA Network and Patterns of Immune Infiltration in Mesothelioma With Bone Metastasis. Front Bioeng Biotechnol 2019;7:257. [Crossref] [PubMed]
- Huang R, Meng T, Chen R, et al. The construction and analysis of tumor-infiltrating immune cell and ceRNA networks in recurrent soft tissue sarcoma. Aging (Albany NY) 2019;11:10116-43. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Kang K, Xie F, Mao J, et al. Significance of Tumor Mutation Burden in Immune Infiltration and Prognosis in Cutaneous Melanoma. Front Oncol 2020;10:573141 [Crossref] [PubMed]
- Zhang J, Lou W. A Key mRNA-miRNA-lncRNA Competing Endogenous RNA Triple Sub-network Linked to Diagnosis and Prognosis of Hepatocellular Carcinoma. Front Oncol 2020;10:340. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Chang Z, Huang R, Fu W, et al. The Construction and Analysis of ceRNA Network and Patterns of Immune Infiltration in Colon Adenocarcinoma Metastasis. Front Cell Dev Biol 2020;8:688. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Sun XY, Yu SZ, Zhang HP, et al. A signature of 33 immune-related gene pairs predicts clinical outcome in hepatocellular carcinoma. Cancer Med 2020;9:2868-78. [Crossref] [PubMed]
- Li R, Qu H, Wang S, et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics 2018;34:2515-7. [Crossref] [PubMed]
- Howlader N, Noone AM, Krapcho M, et al. SEER cancer statistics review, 1975-2016, National Cancer Institute. Bethesda, MD. 2019. https://seer.cancer.gov/csr/based on November 2018 SEER data submission, posted to the SEER web site.
- Alfred Witjes J, Lebret T, Compérat EM, et al. Updated 2016 EAU Guidelines on Muscle-invasive and Metastatic Bladder Cancer. Eur Urol 2017;71:462-75. [Crossref] [PubMed]
- Smith AB, Jaeger B, Pinheiro LC, et al. Impact of bladder cancer on health-related quality of life. BJU Int 2018;121:549-57. [Crossref] [PubMed]
- Zheng X, Wang X, Zheng L, et al. Construction and Analysis of the Tumor-Specific mRNA-miRNA-lncRNA Network in Gastric Cancer. Front Pharmacol 2020;11:1112. [Crossref] [PubMed]
- Ma L, Song G, Li M, et al. Construction and Comprehensive Analysis of a ceRNA Network to Reveal Potential Novel Biomarkers for Triple-Negative Breast Cancer. Cancer Manag Res 2020;12:7061-75. [Crossref] [PubMed]
- Liu S, Song A, Zhou X, et al. ceRNA network development and tumour-infiltrating immune cell analysis of metastatic breast cancer to bone. J Bone Oncol 2020;24:100304 [Crossref] [PubMed]
- Duan S, Wang P, Liu F, et al. Novel immune-risk score of gastric cancer: A molecular prediction model combining the value of immune-risk status and chemosensitivity. Cancer Med 2019;8:2675-85. [Crossref] [PubMed]
- Angell H, Galon J. From the immune contexture to the Immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol 2013;25:261-7. [Crossref] [PubMed]
- Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938-45. [Crossref] [PubMed]
- Huang CM, Huang CS, Hsu TN, et al. Disruption of Cancer Metabolic SREBP1/miR-142-5p Suppresses Epithelial-Mesenchymal Transition and Stemness in Esophageal Carcinoma. Cells 2019;9:7. [Crossref] [PubMed]
- Liu C, Chikina M, Deshpande R, et al. Treg Cells Promote the SREBP1-Dependent Metabolic Fitness of Tumor-Promoting Macrophages via Repression of CD8(+) T Cell-Derived Interferon-γ. Immunity 2019;51:381-97.e6. [Crossref] [PubMed]
- Li J, Ping JL, Ma B, et al. DIP2C expression in breast cancer and its clinical significance. Pathol Res Pract 2017;213:1394-9. [Crossref] [PubMed]
- Mir R, Pradhan SJ, Patil P, et al. Wnt/β-catenin signaling regulated SATB1 promotes colorectal cancer tumorigenesis and progression. Oncogene 2016;35:1679-91. [Crossref] [PubMed]
- Mao LJ, Yang CH, Fan L, et al. SATB1 promotes prostate cancer metastasis by the regulation of epithelial-mesenchymal transition. Biomed Pharmacother 2016;79:1-8. [Crossref] [PubMed]
- Zhai S, Xue J, Wang Z, et al. High expression of special AT-rich sequence binding protein-1 predicts esophageal squamous cell carcinoma relapse and poor prognosis. Oncol Lett 2017;14:7455-60. [Crossref] [PubMed]
- Chen Z, Li Z, Li W, et al. SATB1 Promotes Pancreatic Cancer Growth and Invasion Depending on MYC Activation. Dig Dis Sci 2015;60:3304-17. [Crossref] [PubMed]
- Huang Y, Yang N. MicroRNA-20a-5p inhibits epithelial to mesenchymal transition and invasion of endometrial cancer cells by targeting STAT3. Int J Clin Exp Pathol 2018;11:5715-24. [PubMed]
- Genin M, Clement F, Fattaccioli A, et al. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 2015;15:577. [Crossref] [PubMed]
- Agarbati S, Mascitti M, Paolucci E, et al. Prognostic Relevance of Macrophage Phenotypes in High-grade Oral Tongue Squamous Cell Carcinomas. Appl Immunohistochem Mol Morphol 2021;29:359-65. [Crossref] [PubMed]
- Jamieson T, Clarke M, Steele CW, et al. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J Clin Invest 2012;122:3127-44. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Fridlender ZG, Sun J, Kim S, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell 2009;16:183-94. [Crossref] [PubMed]
- Hayashi T, Sentani K, Oue N, et al. Desmocollin 2 is a new immunohistochemical marker indicative of squamous differentiation in urothelial carcinoma. Histopathology 2011;59:710-21. [Crossref] [PubMed]
- Oldford SA, Marshall JS. Mast cells as targets for immunotherapy of solid tumors. Mol Immunol 2015;63:113-24. [Crossref] [PubMed]
- Fang R, Huang Y, Xie J, et al. Downregulation of miR-29c-3p is associated with a poor prognosis in patients with laryngeal squamous cell carcinoma. Diagn Pathol 2019;14:109. [Crossref] [PubMed]
- Wang Y, Ba HJ, Liu ZC, et al. Prognostic value of immune cell infiltration in bladder cancer: A gene expression-based study. Oncol Lett 2020;20:1677-84. [Crossref] [PubMed]


