
A narrative review of urinary phospholipids: from biochemical aspect towards clinical application
Introduction
Urine is a unique, easily accessible, and noninvasive sample that is widely used for clinical diagnostics, especially in the field of urology. The analysis of urinary components has often been utilized for assessing metabolic or physiological states of patients or healthy subjects (1). Additionally, urine is an attractive matrix for biomarker discovery, presenting rich metabolite categories and high participant compliance (2).
The concept of lipidomics was introduced by Spener et al. (3) and Han and Gross (4) in 2003, which refers to the qualitative and quantitative analysis of global lipids in a given cellular or biological system. In recent years, phospholipids (PLs) have been separately analyzed from other lipids owing to their unique amphipathic properties (hydrophobic fatty acyl tails and hydrophilic head group on the same molecule), diverse bio-functions, and biological relevance in various diseases (5).
In cells, owing to their amphipathic properties, PLs are the main components of bilayer membranes of the cell and intracellular organelles, providing cellular integrity (5). Additionally, PLs act as key members in cellular signaling or sources of energy for various cellular processes, including homeostasis, cell migration, apoptosis, autophagy, and post-translational modifications (6).
Alterations in the composition of various PLs in cells, tissues, and body fluids, including urine, are associated with numerous urologic and genitourinary diseases and pathophysiological conditions such as genitourinary (prostate, bladder, and kidney) cancers (6), kidney stones (7), inflammatory disorders (8), and sperm quality (9).
Owing to the extremely low concentrations of PLs and high levels of contamination in urine, an effective extraction method, as well as a sensitive detection method for urinary PLs remain crucial for clinical application. Traditional liquid-liquid extraction (LLE) methods, including Folch (10), Bligh-Dyer (11), butanol-methanol (BUME) (12,13), methyl-tert-butyl ether (MTBE) (14), and hexane-isopropanol (HIP) (15), were applied and compared for urinary PLs and lysophospholipids (LPLs) in our previous study (16). Furthermore, diverse analytical methods such as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (17), electrospray ionization mass spectrometry (ESI-MS) (18), liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) (19), and nuclear magnetic resonance spectroscopy (NMR) (20) have been utilized for detecting urinary PLs and LPLs in previous reports.
In this review, we focus on the chemical, physical, and biological aspects of urinary PLs and LPLs, as well as their clinical applications in genitourinary diseases. We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1263).
Methods
This narrative review designed to collect and review published articles and literatures on the biochemical aspects of PLs and LPLs and the clinical application of urinary PLs and LPLs. A search was carried out on the databases including PubMed, Web of Science and Google Scholar by MeSH-compliant keywords including phospholipid(s), LPL(s), biochemical property, urine, biomarkers, and genitourinary diseases. All related original research studies, letters to the editor, reviews and ebooks written in English and published from January 1950 to August 2020 were screened and checked.
Discussion
Chemical and physical aspects of PLs and LPLs
Chemical structure
PL is composed of a glycerol backbone, two fatty acids, and a phosphate group. The chemical structures of the most common PLs are presented in Figure 1. In PLs or LPLs, a glycerol molecule forms the backbone of the whole structure (shown in Figure 1). Each carbon in the glycerol molecule is numbered using the “stereospecific numbering” (sn) system. The hydroxyl groups at the sn-1 and sn-2 positions are esterified with two fatty acids (shown in Figure 1), and the hydroxyl group at sn-3 is esterified with a phosphate group to form a head group. Variations of head groups (shown in the dashed box in Figure 1) extend the subcategories of PLs, resulting in phosphatidic acid (PA, with hydrogen), phosphatidylcholine (PC, with choline), phosphatidylethanolamine (PE, with ethanolamine), phosphatidylserine (PS, with serine), phosphatidylglycerol (PG, with glycerol), and phosphatidylinositol (PI, with inositol). Within each sub-category, the length and saturation of hydrophobic fatty acid chains lead to further characterization of glycerol-PLs. For example, dilauroyl PLs (12 carbon atoms in each fatty acyl chain), dimyristoyl PLs (14 carbon atoms in each fatty acyl chain), or dipalmitoyl PLs (16 carbon atoms in each fatty acyl chain) are referred to based on the length of fatty acyl chains. Dioleoyl PLs (containing one double bond in each fatty acyl chain) and distearoyl PLs (containing two double bonds in each fatty acyl chain) are characterized by the saturation condition of fatty acyl chains. If an acyl group is connected to the glycerol backbone (shown in the left bottom of Figure 1), it is termed a LPL, which can be catalyzed by PLs in the cell. Variations of LPL head groups is similar to PLs, resulting in subclasses such as lysophosphatidic acid (LPA), lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), lysophosphatidylserine (LPS), lysophosphatidylglycerol (LPG), and lysophosphatidylinositol (LPI) by connecting the phosphate with hydrogen, choline, ethanolamine, serine, glycerol, and inositol, respectively.
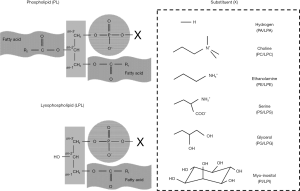
Physical properties
Solubility
As lipids, PLs and LPLs are poorly soluble in water and other polar solvents but are soluble in organic solvents such as chloroform (CHCl3), methanol (MeOH), hexane, ethanol, isopropanol, or in mixtures these solvents. This property allows PLs and LPLs to be easily separated and extracted from aqueous solution (e.g., urine) by the addition of organic solvents. For example, in our previously optimized extraction method for urinary PLs and LPLs (16), 5 mL of an organic solvent mixture containing CHCl3/MeOH (1:1, v/v) was added to 1 mL of urine, followed by the addition of 1.25 mL of water. The volume ratio of CHCl3, MeOH, and water in the final solution was approximately 1:1:0.9, achieving an optimal condition for transferring PLs from the aqueous to the solvent phase. Accordingly, lipids, including PLs, are isolated and purified from urine.
Electrolysis
Notably, all PLs vary from no net charge (PC/LPC and PE/LPE, also named zwitterionic PLs or LPLs) to anions (PA/LPA, PS/LPS, PG/LPG, and PI/LPI), with none possessing a net positive charge under physiological pH. Therefore, anionic PLs can provide negative charge densities to the cellular/organelle membranes, necessary for recruitment and interactions with ions and proteins (21). At physiological pH, zwitterionic PLs (PC/LPC and PE/LPE) contain both a negative and positive charge group. A pH change in the aqueous environment causes protonation/deprotonation of the PL head groups, greatly impacting the chemical and physical properties (e.g., hydration, swelling, phase transitions, and the overall polarity) of the PL molecule. The altered PL properties will further influence the interactions with ions and solutes such as peptides and proteins, causing a downstream reaction in the cell/organelle. If the pH of the solution is equal to the isoelectric point of PLs, their physicochemical behavior will resemble that of nonionic lipids (e.g., mono- and digalactosyldiacylglycerols). Furthermore, a change in pH may influence the hydrolytic stability of PLs, which are more rapidly hydrolyzed at high pH than at low pH.
Amphiphilicity
Although poorly water-soluble, as described in the previous section, PLs are readily dispersible in water owing to a feature termed ‘amphiphilicity’. The word ‘amphi’, derived from Greek, means ‘both’. In the case of PLs in water, all PL molecules are composed of two parts: a hydrophilic part (the phosphate group) and hydrophobic part (fatty acid chains) (22). The amphiphilic property is essential for the spontaneous formation of the lipid bilayer structure in aqueous environments. The hydrophilic phosphate head group faces water at each surface of the bilayer, whereas the hydrophobic fatty acid tails remain shielded from water in the interior (23). This is the fundamental structure of the cell and organelle membrane, which is further discussed in a later section.
Biological Functions of PLs and LPLs
As described above, physical, and chemical diversities confer PLs versatile bio-functions in cells. Cumulative studies have indicated the three main roles of PLs, including as major components of cellular and organelle membranes, signaling mediators in various cellular processes, and members of energy storage (24). Table 1 summarizes the main bio-functions of PLs and LPLs described in previous studies.

Full table
Membrane
For PLs, the first illustrated biological function is the formation of cell and organelle membranes, which help provide cellular integrity and separation of subcellular compartments, which are fundamental requirements for all cellular functions. PLs can form an ordered bilayer structure owing to their amphiphilic nature (described above). The hydrophobic fatty acyl tails of PLs line up and face each other, whereas the hydrophilic head group faces both sides of the membrane. This membrane is described as a ‘fluid mosaic model’ (23), which is semi-fluid and partially permeable, with embedded proteins in the PL matrix.
Among the different subcategories of PLs, PC has demonstrated the highest content in animal membranes (25). It is ideally suited to this role as the width of its polar head group is approximately identical to the sum of its apolar fatty acyl tails (26). Moreover, PC is a zwitterion with a formal positive and negative charge, lacking a net charge. Owing to these two physical properties, PC is deemed relatively stable in membranes, both physically and chemically. PE is the second most prevalent PL in the human body and is particularly rich in nervous tissues. As another zwitterionic PL, the primary role of PE is as a structural lipid in the cell membrane (25). For PE, the head group is small and poorly hydrated, imparting a pyramidal shape, with a wide base (fatty acyl chains) and a narrow head. Owing to its amine-containing head group, PEs are also chemically active and can form hydrogen bonds with other neighboring polar groups, thus impacting the membrane structure (27).
To considerably lower levels when compared with PC and PE, other anionic PLs (i.e., PA, PS, PG, PI, and those LPLs) also play important roles, with additional functions in the cell membranes. For example, the net charge of PA at physiological pH is only 1.5, and such a strong anion contributes negative charge density to the membrane, which is necessary for membrane vesicle fission, as well as the activities of some membrane proteins (30). Like the anionic role of PA, PS, PI, and PG additionally act as important anions at physiological pH and thus contribute to the negative charge density of lipid bilayer membranes. Indeed, they possess additional functions in cell signaling and regulation in addition to structural lipids in the cells; this will be discussed in the next section.
Cell signaling
Beyond their role as a fundamental structure of lipid bilayers, PLs participate in cellular processes such as signaling. Owing to their transmembrane location and diversity in structural modification, PLs play crucial roles in various cellular functions by directly regulating enzymes or acting as precursors of signaling molecules. Herein, we briefly introduce several widely investigated PLs demonstrating important biological functions in cell signaling.
Phosphoinositides
Phosphoinositides, the phosphorylated derivatives of PI, play a central role in cell signaling and regulation (50,51). Frequently, PI is phosphorylated at various positions on the inositol chain. The D-3, D-4, and/or D-5 positions of the hydroxyl groups of the inositol ring result in the formation of seven distinct phosphoinositide species, including phosphatidylinositol 3-phosphate (PI3P), PI4P, PI5P, phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2], PI(3,5)P2, PI(4,5)P2, and phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3]. Phosphoinositides are localized on inner plasma membranes, including various organelle (endosomal, Golgi, and nuclear) membranes (mainly monophosphates) and inner plasma membranes (mainly di- and triphosphates), tightly regulated by a network of kinases and phosphatases. Phosphoinositides regulate cell processes through direct interaction and modeling of membrane proteins (such as ion channels) or recruitment of cytosolic proteins to membranes, and activating downstream pathways [such as protein kinase B, also known as Akt, can be recruited and activated by PI(3,4)P2 or PI(3,4,5)P3], thus regulating cellular processes such as proliferation, apoptosis, and migration. Phosphoinositide signaling affects various cellular processes, including proliferation (52), survival (53), migration (54), cytoskeletal rearrangement (55), vesicle trafficking (56), membrane dynamics (57), and apoptosis (58).
Phosphatidic acid
As the simplest PL with relatively low abundance (covering only 1–4% of the total PLs), PA is reportedly known as an intermediate for the biosynthesis of other PLs following dephosphorylation to diacylglycerol (DAG). In recent years, PA has been recognized as a signaling messenger (31). In cells, PA levels are dynamic and controlled (i.e., synthesis and catabolism) by several enzymes, including phospholipases (e.g., phospholipase A and D), lipid kinases (e.g., PA kinase, DAG kinase), and phosphatases (e.g., lipid phosphate phosphatase). The cellular level of PA is highly increased under stress conditions such as injury, freezing, and infection (31). Although yet to be thoroughly elucidated, PA reportedly interacts with effector proteins in the “electrostatic/hydrophobic switch mechanism” (32) and “electrostatic/hydrogen-bond switch mechanism” (33), performing its role as a cellular messenger. PA recruits effector proteins to the membrane and regulates the activities of proteins in cellular pathways. Some effector proteins known to interact with PA are Son of sevenless (Sos) (34), kinase Raf-1 (35), mammalian target of rapamycin (mTOR) (36), Gardner-Rasheed feline sarcoma viral (FGR) (37), and sphingosine kinase (38), which are involved in multiple signaling pathways such as Akt (39), Hippo (40), and vascular endothelial growth factor (VEGF) (41). These PA-protein interactions are involved in a wide range of cellular processes, including cell proliferation, apoptosis, vesicular trafficking and secretion, and cytoskeletal organization (42), as well as pathophysiological conditions like inflammation (43), oncogenesis, and metastasis (44). Importantly, PA is known to play an important role in spermatogenesis by facilitating the generation of piwi-interacting RNA (piRNA), a critical event in germline development (45).
Phosphatidylserine (PS)
Reportedly, PS is involved in cell-cell recognition and communication processes during apoptosis (46). In contrast to other PLs, PS is located almost exclusively in the membrane inner leaflet, which is transported and maintained by ATP-dependent amino-PL translocases (also known as flippases) (46). Extracellularly, PS is involved in platelet activation (46) and also serves as an “eat me” signal for the clearance of apoptotic cells (47). Intracellularly, several pathways depend on PS. For example, the PS-specific binding of Raf kinases is involved in the activation of mitogen-activated protein kinase (MAPK) signaling (28).
Phosphatidylethanolamine (PE)
One important and attractive function of PE in cells is its involvement in autophagosome biogenesis during autophagy. In the autophagic process of mammalian cells, PE attaches to microtubule-associated protein light chain 3 (LC3), which functions as an anchor to autophagosomal membranes for LC3 (29). Reportedly, increased intracellular PE levels significantly increased autophagic flux (80). Furthermore, PEs possess other diverse cellular functions, including serving as a precursor for PC and PS synthesis, a determinant of membrane protein topology, and promoting the membrane fusion of cells and organelles by accumulation on the external leaflet (29).
Phosphatidylglycerol (PG)
PG is the second most abundant lipid present in the lung surfactant (LS) of mammals, demonstrating a fluidizing effect on LS. In fetuses, PG levels in the LS are used as indicators of lung maturity, and a lack of PG causes respiratory distress syndrome (RDS) (81). However, the role of PG in human cell signaling remains limited. Previous in vitro studies have indicated that PG is involved in the activation of RNA synthesis (48). In cells, the lack of PG leads to defective cell wall synthesis, and PG is required in protein kinase C (PKC) signaling (49).
LPLs as lipid mediators
LPLs, particularly LPAs, are important bioactive lipids in the human body and can act as hormones in the blood by combining with a group of specific G protein-coupled receptors (GPCRs) expressed on the surfaces of target cells (69). As the most investigated LPL, LPA is an extracellular mediator catabolized from substrate LPC and PA. Reportedly, six GPCRs were determined for LPA ligand recognition. On binding to these receptors, LPA activates or inhibits downstream secondary messengers, including MAPK, phosphoinositol 3-kinase (PI3K), Rho, Ras, DAG, adenylyl cyclase (AC) inositol 1,4,5-trisphosphate (IP3), and Ca2+, in various pathways (66). LPA signaling plays key roles in tumor migration and invasion, smooth muscle contraction, vascular development, inflammation, endothelial integrity, neurogenesis, implantation of fertilized eggs, and lymphocyte homing (67-69). Although not as popular as LPA, LPC, which is the most abundant LPL in blood, has attracted increasing attention as a key lipid factor in various pathophysiological conditions such as inflammation, fatty acid oxidation, endothelial activation, and atherogenesis (59-62). Currently, three LPS GPCRs have been identified, and their roles have attracted attention in the immunomodulation (69). In previously reported studies, LPS signaling was involved in platelet activation, inflammation, and the pathogenesis of atherosclerotic diseases (69-71). LPE was found to play important roles in neuronal differentiation via the activation of MAPK (63). In addition, LPE can increase intracellular calcium, thus facilitating the migration and invasion of ovarian and breast cancer (BC) cells (64,65). Like LPE, LPG has been found to stimulate calcium signaling and increase intracellular calcium in ovarian cancer cells (72). Moreover, this ability of LPG to increase intracellular calcium can inhibit chemokine-induced migration and interleukin-1β production in human phagocytes (73). The first reported biological role of LPI was the stimulatory effect of insulin release by pancreatic islets (74,75). Since then, a growing number of LPI bio-functions/roles have been revealed, including as a potent mitogenic factor, and in fat deposition and wound healing (76-79).
Although current studies mainly used serum or plasma as sources of LPLs biomarkers, a previous review indicated that urine could be utilized as another promising sample for LPL assays as urine requires no burden on the subject. Furthermore, LPLs in the urine are relatively more stable than in blood, which comprises enzymes that introduce undesired and artificial effects, thus facilitating the in vitro manipulation of LPLs (69).
Cellular bioenergetics
Moreover, PLs regulate the architecture, biogenesis, and degradation of lipid droplets (LDs), which play critical roles in the energy homeostasis of cells. LDs consist of a core of neutral lipids wrapped by a PL monolayer with integral/peripheral proteins. This lipid-rich cellular organelle is believed to play a central role in the storage and supply of energy in nearly all cell types (82). LDs are highly dynamic organelles that alternate between periods of formation and consumption (83). The LD monolayer not only provides a barrier between LD and the cytoplasm, but is also involved in the communication between LDs and other cellular organelles (e.g., endoplasmic reticulum, mitochondria, and lysosomes) (84). This communication determines the formation and mobilization of LDs in conditions of surplus nutrients or starvation. Furthermore, a recent study indicated that PLs also determine the positions of LDs; PLs with large positive molecular curvature (such as LPLs) facilitate the emergence of LDs from the endoplasmic reticulum (ER), while negative molecular curvature PLs (such as PE) favor the embedding of LDs in the ER (85).
Methods for measuring urinary PL and LPLs
Owing to their extremely low concentration and high diversity in structures, comprehensive analysis and quantification of PLs and LPLs present a considerable challenge. For urinary PL and LPL analysis, the general workflow mainly includes lipid extraction from the urine, as well as qualitative and quantitative analysis by diverse approaches. A wide range of technologies such as MS, chromatography including high-performance liquid chromatography (HPLC) (86), thin-layer chromatography (TLC) (87), and NMR spectroscopy (88) are commonly utilized for the detection of PLs. Among these, the MS-based method is considered the primary choice for the identification and quantification of PLs and LPLs in the urine. Thus, we mainly focused on the application of MS for the analysis of urinary PLs and LPLs. A common workflow for the MS-based urinary PL and LPL study is shown in Figure 2, which includes three main steps of sample preparation (i.e., collection of urine samples and extraction of urinary lipids), data acquisition by MS analysis, and MS data processing by bioinformatics for the screening potential biomarkers. We summarized urinary PLs and LPLs detected in our laboratory using MALDI-TOF/MS in both positive and negative modes.
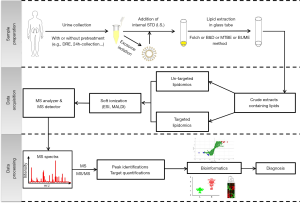
Extraction
The extraction of lipids from biological samples such as urine is the first step in any lipid analysis method. Prior to analysis, enrichment and purification of PLs from urine are necessary owing to the considerably low lipid concentration when compared with the high abundance of salts and other charged compounds such as amino acids (17).
LLE using a mixture of organic solvents is the most commonly used method to solubilize and separate lipids from the matrix (e.g., tissues and fluids), which are largely insoluble in these organic solvents. Furthermore, solvent extraction can help to minimize the oxidation of unsaturated fatty acyl chains in PLs (89). The most popular choice of organic solvents is a mixture of CHCl3 and MeOH, applied in two methods that were developed by Folch et al. (10) in 1951 and Bligh and Dyer (11) in 1959. Although introduced more than 60 years ago, these two methods remain widely utilized and play a fundamental role in several other modified approaches. The Folch method uses CHCl3 and MeOH in a 2:1 ratio, which is suitable for lipid extraction from tissues, whereas the Bligh and Dyer (B&D) method uses CHCl3 and MeOH in a 1:1 ratio and is commonly designed for lipid extraction from fluids with a large amount of endogenous water. These CHCl3/MeOH based methods have been widely applied in urinary lipid studies. For instance, Min et al. used the Folch method to extract urinary PLs and LPLs from lyophilized urine powder (90); Rockwell et al. used the B&D method for liquid urine samples (91); Skotland et al. used a modified Folch method for extracting lipids from urinary exosomes (92). Although demonstrating high PL recovery rates, the high toxicity of CHCl3 has promoted the exploration of alternative organic solvents/methods for achieving the goal of a “green extraction process”. Matyash et al. developed a mixture of methyl tertiary butyl ether (MTBE)/MeOH for replacing CHCl3 lipids and revealed similar or even better recoveries for species of most major lipid classes when compared with the Folch or B&D methods in E. coli, mouse brain tissue, human plasma, and C. elegans embryos (14). Additionally, this method has been applied in lipid extractions from urine (93) and urinary exosome (94) in later studies. LLE using other solvents such as 1-butanol/MeOH (BUME) (13) has been performed for the evaluation of urinary PLs and LPLs (95). To optimize the process of the urinary PL and LPL workflow and select one suitable lipid extraction for MALDI based urinary PL evaluation, our group tested six different lipid extraction methods, including the Folch, B&D, acidified B&D, MTBE, BUME, and hexane/isopropanol (HIP). We observed that using MALDI-TOF/MS as a detection system with 9-aminoacridine as the matrix, the acidified B&D method showed excellent recovery rates for both positively and negatively charged PLs in urine (16). This method may offer a rapid and comprehensive screening for PLs and LPLs in urine samples.
Diverse analytical technologies
After the extraction of urinary lipids, the crude extracts are analyzed to exploit qualitative and quantitative information regarding the lipid class or individual molecular species using diverse techniques. Commonly used methods involve NMR, which is extremely suitable for PL identification but requires a relatively high sample content and expensive instrumentation, basic chromatography that simply separates lipid classes but cannot isolate individual lipid species, and MS-based lipidomics with high sensitivity, resolution, and throughput, which has become the primary choice for identification and quantification of urinary PLs and LPLs.
Chromatography-based phospholipidomics
Chromatography is a technique generally used for separating complex mixtures into simpler groups. There are three main basic methods, including TLC, HPLC, and gas chromatography (GC), commonly used for lipid studies. Among them, TLC (87) or HPLC (86) are more commonly utilized than GC for the detection of PLs as GC is best suited for volatile lipids that are thermally stable and have a molecular weight less than 500 Da (87).
TLC separates a sample mixture by migrating the organic solvents on a silica-coated glass plate. Samples deposited on a solid surface are separated based on affinity to the solvent, and molecules that are more preferred to the solvent (e.g., lipids) can move further up on the plate than those that prefer the solvent. The separated lipids can be visualized or further analyzed by MS. TLC can further resolve lipids by class, or by the length or saturation of acyl chains. The advantages of TLC include a simple and fast experimental process and inexpensive instrumentation, rendering it easy to implement in laboratories. However, the main disadvantage of TLC is the limited resolution that fails to separate individual lipid species, and thus demonstrating limited application in advanced lipid separation. Using TLC, Boonla et al. separated and identified major lipid classes in total lipid extracts obtained from urine and stones collected from patients with nephrolithiasis (96). In lipid research, a more important application of TLC is to combine TLC separation with MS analysis. The identification of PL species can be performed by coupling MS on the TLC plate (97).
The HPLC system separates each component in a mixture by pumping a liquid solvent containing the sample mixture (mobile phase) that flows through a solid column (stationary phase). All components within the sample mixture possess different molecular weights and slightly different affinities to the stationary and mobile phases, thus demonstrating different flow speeds, and finally leading to their separation while flowing through the column. HPLC is a versatile chromatographic technique that enables the separation of most lipids and PL classes, using a combination of numerous columns and solvents. Importantly, the coupling of HPLC with MS has dramatically increased the utility of HPLC (will be discussed later). Several previous studies have used the HPLC-MS technique for the sensitive detection of urinary PLs and LPLs (94,98,99).
NMR spectroscopy-based phospholipidomics
In addition to chromatography, different spectroscopic techniques have been used for lipid analysis. Among these, NMR spectroscopy is capable of providing the most detailed information regarding the lipid composition of a mixture. One convenient method in PL research is 31P NMR—when PL amounts are in the 100 µg range (20). By using a suitable solvent system, highly resolved 31P NMR spectra can be obtained in a few minutes, even from complex crude mixtures. These spectra allow the differentiation of all major PL classes and clear differentiation of the fatty acyl residues in PLs (100) (i.e., saturated, moderately unsaturated, and highly unsaturated lipids can be differentiated). Additionally, a positional analysis of the fatty acyl residues is possible using this technique (101). For instance, isomers of LPC (sn1- and sn2-LPC) can be differentiated utilizing their different chemical shifts. However, NMR requires larger amounts of PLs than chromatography or MS (20). Another disadvantage is the high price of 31P NMR, limiting its application. Schiller and coworkers developed protocols for combining 31P NMR with MALDI-TOF/MS, revealing both excellent sensitivity detection, as well as precise identification and quantification of PLs (102).
Mass spectrometry-based phospholipidomics
As mentioned above, MS plays a central role in the analysis of PLs, either combined with chromatography or NMR. Typically, a mass spectrometer consists of three main parts: an ion source that generates ionized/vaporized analytes, a mass analyzer system that separates the ions according to their mass to charge (m/z) ratio, and a detector that records the signal intensities of different ionized molecules with different m/z values. By plotting the m/z value (x-axis) and signal intensity (y-axis), a mass spectrum can be displayed. Mass spectrometers can be classified into different types based on different types of ion sources and mass analyzers. Within the scope of PL studies, soft ionization methods involving ESI and MALDI are used to avoid molecular fragmentation. Mass analyzers used for PL analysis mainly include linear ion trap (quadrupole), triple quadrupole, time-of-flight (TOF), Orbitrap, and a hybrid of these techniques (103). The analytical chemistry background of these techniques is beyond the scope of this review. Thus, we focused on the introduction of established MS approaches for urinary PL studies—targeted and untargeted lipidomics using ESI-MS or MALDI MS platform.
Typically, in the ESI platform, according to whether prior chromatographic separation (usually HPLC) of the lipid mixture has been performed before the MS analysis, the ESI lipidomic approach for urinary PL evaluation can be divided into two strategies: untargeted lipidomics (also termed shotgun lipidomics) and targeted lipidomics (LC-ESI-MS/MS). Direct-infusion ESI untargeted lipidomics was developed by Han and Gross approximately 20 years ago and is currently one of the most widely used methods in PL and LPL studies (104). This was initially performed using direct-infusion ESI coupled with a triple quadrupole (Q) analyzer; however, presently, several newly developed technologies, including chip-based nanoLC-ESI, Q-TOF, Fourier transform–ion cyclotron resonance (FT-ICR), and orbitrap analyzers, have dramatically enriched the choice of available methods that enhance mass resolution and detection limits under various conditions (105). As the goal of untargeted lipidomics is to identify and quantify the maximum number of lipid molecular species in a single run, the ability to measure all detectable lipids and quantify targets with just one non-endogenous internal standard for each lipid class remain the main advantages of this method. These advantages establish untargeted lipidomics as a promising strategy to screen PL biomarkers in complex biological samples such as urine. However, untargeted lipidomics is prone to the ion suppression effect observed with minor lipid molecules with poor ionization properties mediated by major lipid molecules with strong ionization properties. Another disadvantage is the failure or difficulty in separating isobaric and isomeric lipid molecules when analyzing complex samples.
Compared with the untargeted lipidomics strategy, targeted lipidomics utilizes a pre-separation before MS analysis using HPLC instruments. As mentioned above, this combined technology overcomes some drawbacks presented by untargeted lipidomics, permits effective separation of PL isomers and isobars, and reduces the ion suppression effects. There are three types of HPLC columns used for PL investigations: reversed-phase LC (RPLC), which separates PLs according to the length of fatty acid chains and the number of double bonds; normal-phase LC (NPLC), which separates PLs mainly based on chemical structures of the head groups; hydrophilic interaction chromatography (HILIC), a variant of NPLC that enables the effective separation of small polar compounds in polar stationary phases (24). The main disadvantages of targeted lipidomics include the complexity in choosing a suitable column for different target molecules and the need for several internal standards per lipid class as the species may spread across a wide retention time window.
Although the application of MALDI for lipid analysis is not as extensive as that of the ESI platform, interest in MALDI has continued to increase owing to the progress in instrumentation, as well as its obvious advantages of fast and highly sensitive data acquisition. A previous opinion regarding the disadvantage of MALDI MS analysis is the weak quantitative ability owing to uneven molecular distribution in the solid analyte-matrix crystal on the MALDI plate. However, this might apply to polar biomolecules (such as proteins), but not PLs (106). This is because both PLs and the matrix are readily solved in organic solvents, and by selecting proper solvents for lipid and matrix, homogeneity of matrix/analyte crystallization can be achieved, offering reliable quantification even in a short analysis (107-109). The other main drawbacks of MALDI lipid analysis, such as matrix overlap of the analytes, could be resolved by the progress in matrix science (110), which has conferred MALDI a promising method in PL investigation, including urinary PLs and LPLs biomarker screening.
Similar to the ESI platform, MALDI can be placed in two strategies based on whether a chromatographic separation is performed prior to MS analysis. As MALDI is mainly recognized as a fast tool for sensitive molecule detection, the major application of this technique is the direct analysis of crude lipid extraction. Matrices applied in lipid research include 2,5-dihydroxybenzoic acid (DHB), α-cyanocinnamic acid (CHCA), and 9-aminoacridine (9-AA), or a mixture of selected options at an appropriate concentration ratio (110).
In our group, we achieved the sensitive detection of urinary PLs and LPLs in the femtomole grades using 9-AA as the matrix (16). We detected urinary PC, LPC, and SM in the positive ionization mode (summarized in Table 2), while analyzing PA, PI, PG, PE, and PS in the negative ionization mode (summarized in Table 3). The relative quantification of the urinary PLs and LPLs has been achieved by the addition of internal standards before lipid extraction. Only two internal/ionization standard species (i.e., non-endogenous commercial PC for the positive ionization mode and non-endogenous commercial PG for the negative ionization mode) can offer a reliable quantification of the bio-PLs by careful optimization of the matrix, solvent systems, and ionization modes (16). Herein, we defined and named this quantification method as a quantitative shotgun MALDI (qShot MALDI) lipidomics, which relies on non-endogenous internal/ionization standard species to rapidly evaluate the relative PL concentration in crude extracts.
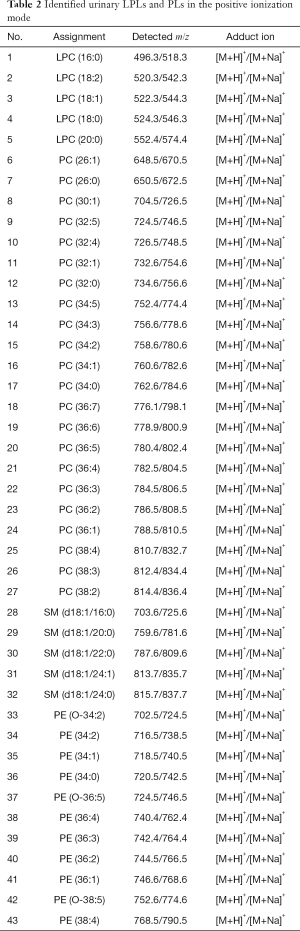
Full table
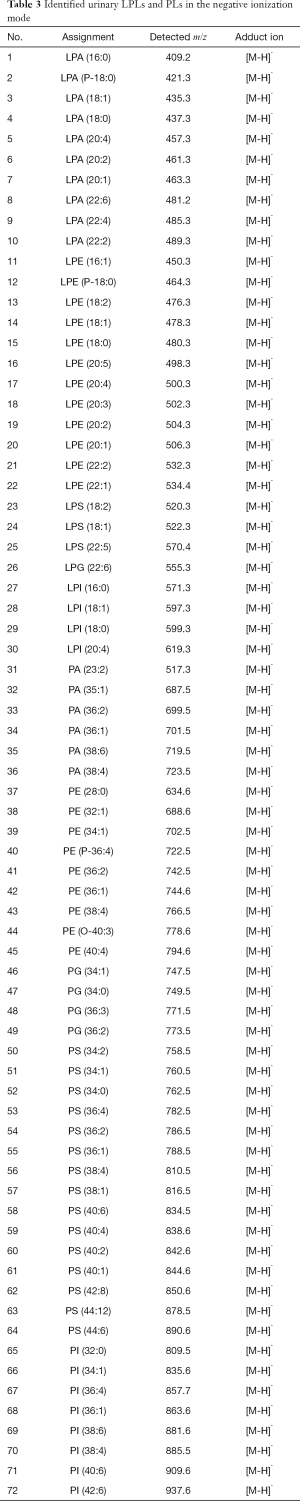
Full table
Furthermore, the pre-separation of lipid classes or species has been achieved by the ingenious combination of MALDI with chromatography in previous studies (111-113). In contrast to ESI, MALDI is usually linked with TLC plates but not LC separation because MALDI is mostly performed under vacuum and is not suitable for combination with LC. Utilizing TLC separation, PLs can be separated and subsequently analyzed by MALDI-TOF/MS, and PLs even less than 1% of the total extract can be sensitively and undoubtedly detected without overlap with other molecules, and matrix peaks can be obtained (111). Additionally, this method has been validated as a reliable system as the relative composition of PLs in the mixture remained constant and only marginal changes were observed in the fatty acyl composition during TLC separation (112).
Clinical application of urinary PLs and LPLs
Owing to the ease of sample collection and rapid development of analytical techniques such as MS, urine has become an attractive source of biomarkers for clinical diagnostics and/or prognostics in various diseases, including genitourinary cancers and renal diseases. The clinical applications of urinary PL and LPLs as potential diagnostic markers for cancerous and benign diseases are summarized in Table 4.
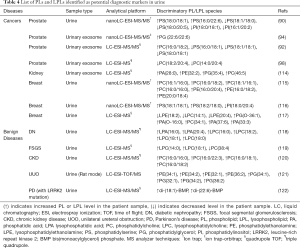
Full table
PLs under normal physiological
Urine contains trace amounts of PLs under normal circumstances. Although present at markedly low concentrations, urinary species are quite variable. The Human Urine Metabolome Database (https://urinemetabolome.ca/) has collected information regarding ~3,100 small molecule metabolites found in human urine, which includes approximately 600 PLs and LPLs. Furthermore, several previous studies reported interesting information about urinary PLs and LPLs under normal conditions. Using the nanoflow liquid chromatography-electrospray ionization tandem MS (nanoLC-ESI-MS/MS) approach, Kim et al. performed a comprehensive analysis of human urinary PLs and identified 75 PL species including 22 PCs, 14 PEs, 15 PIs, 13 PSs, 7 PAs, and 4 PGs in healthy individuals (19). In another study that focused on normal urinary PL secretion (123), lipidomic profiles were quantified in urine samples collected from 16 healthy individuals (including 8 men and 8 women) at 5 fractions over 24 h. Using the shotgun lipidomic approach, six subclasses of PLs (PC, PE, PE-O, PI, PG, and PS) were detected, and a sexual dimorphism between men and women was observed mainly for PC and PE species. In both men and women, PLs were highly correlated with the excretion of urinary creatinine and albumin.
PLs as versatile cancer biomarkers
Altered PL profiles in urine samples have been used to identify specific biomarkers and/or evaluate anticancer treatment in various cancer types, including genitourinary (prostate, bladder, and kidney) and BC.
Genitourinary cancers
As the most concerning disease in urology, urologic malignancies have attracted major investigations within the scope of urinary PL and LPL biomarker screening. Several researchers have selected urinary PLs and LPLs as sources of cancer biomarker candidates because of their noninvasive availability. Min et al. performed qualitative and quantitative profiling of urinary PLs and LPLs using nanoLC-ESI-MS/MS in urine samples collected from 9 prostate cancer (PCa) patients, compared with 10 healthy control individuals. In total, 70 PLs, including 21 PCs, 11 PEs, 17 PSs, 11 PIs, 7 PAs, and 3 PGs were identified (90). Among them, two PS species [PS (18:0/18:1) and PS (16:0/22:6)] showed significantly higher concentrations, while the other two PS [PS (18:1/18:0) and PS (18:0/20:5)] and two PI species [PI (18:0/18:1) and PI (16:1/20:2)] presented significantly lower concentrations in the PCa patient group when compared with the healthy control group. Yang et al. investigated the size distributions, as well as the lipid compositions of urinary exosomes isolated from pooled urine samples from four PCa patients and healthy controls each. Notably, the PCa group demonstrated an increased exosome size. Moreover, 286 lipids, including 118 PL species from urinary exosomes, were identified and quantified. The lipidomic data revealed a general increase in PL amounts in smaller-sized exosomes isolated from PCa patient samples. Among them, PG (22:6/22:6) was approximately 7.5 times higher in the cancer group (94). Additionally, using urinary exosomes, another group of researchers (92) performed high-throughput MS quantitative lipidomic analysis and identified 107 lipid species including 36 PLs. On comparing 15 PCa patients and 13 healthy controls, urinary exosome-derived PC (16:0/18:2) was significantly higher in the PCa group, whereas 3 PS species [PS (18:0/18:1), PS (16:0/18:1) and PS (18:1/18:1)] revealed significantly lower concentrations in the same group. The combination of several PL molecules showed a clear separation between cancer patients and healthy controls, which might be used as a diagnostic index. Clos-Garcia et al. (98) performed a targeted metabolomics analysis of urinary extracellular vesicles (EVs) and compared results between 31 PCa patients and 14 benign prostate hyperplasia (BPH) controls. These results showed decreased levels of PCs [e.g., PC (18:2/20:4), PC (14:0/20:4)] in the PCa group when compared with the BPH group.
In contrast to PCa, urinary biomarker studies of bladder or kidney cancer are limited, although urine is considered a good source for screening specific PL biomarkers in these two diseases. A previous study (114), which utilized a microLC-Q-TOF-MS platform for lipidomic profiling of urinary exosome, was performed using urine samples from 8 renal cell carcinoma (RCC) patients and compared with 8 healthy controls. The results showed that some PL species such as PA (28:0), PE (32:2), PG (35:4), and PC (46:5) were only detected in the RCC group, and could be applied as potential biomarkers for RCC.
Other cancer types
To the best of our knowledge, except in genitourinary cancers, other applications of urinary PLs and LPLs as specific biomarker candidates are mainly in BC. Kim and coworkers (115) qualitatively and quantitatively analyzed urinary PCs and PEs extracted from 5 patients with BC (both before and after surgery) and 5 non-cancer controls. In total, 21 PCs and 12 PEs were identified, 4 urinary PC species [PC (16:1/16:0), PC (16:0/18:2), PC (18:1/16:1), and PC (16:0/16:0)] increased by over two-fold in patients with BC, and these PC species decreased to normal or undetectable levels after surgery. Similarly, in PE, some species, including PE (16:0/20:4), PE (16:0/18:2), and PE (20:0/18:4), were prominently decreased after surgery.
In another study, the same group (116) focused on four other types of PLs (PA, PS, PG, and PI) for screening BC biomarker candidates. Overall, 34 urinary PLs (including 6 PAs, 12 PSs, 4 PGs, and 12 PIs) were quantitatively examined in urine samples collected from 5 patients with BC (both before and after surgery) and 5 non-cancer controls. Among them, two PS species [PS (18:1/18:1) and PS (18:2/18:0)] showed significantly higher concentrations in the BC patient group than in the control group, and their concentrations were found to be reduced to normal levels after surgery. Moreover, one PI molecule [PI (18:0/20:4)] showed a significantly lower concentration in the BC group, decreasing even further after surgery. Cala et al. (117) evaluated urinary lipid alterations in Hispanic Colombian women diagnosed with BC and observed that some LPLs, including LPE (18:2), LPC (14:1), and LPE (20:4), were decreased, whereas PLs such as PG (O-36:1), PA (O-16:0), PC (34:1), PA (37:6), and PA (33:3) were increased in women with BC than in healthy controls.
PLs in benign diseases
Some urinary LPA and LPC species, which are known to promote renal inflammation, were evaluated in urine samples obtained from 41 type II diabetic patients with nephropathy symptoms and 41 type II diabetic controls without nephropathy symptoms (118). The results indicated that the concentrations of six urinary LPA and LPC species [LPA (16:0), LPA (20:4), LPC (16:0), LPC (18:2), LPC (18:1), and LPC (18:0)] were significantly higher in diabetic nephropathy cases than in the non-nephropathy controls (118). In another study investigating renal disease (119), urinary lipid profiles were determined in patients with focal segmental glomerulosclerosis (FSGS) and healthy controls. The results showed an increased concentration of LPC (14:0) and LPC (18:1) and decreased PC (38:4) in patients with FSGS. These species could be used as biomarkers for the diagnosis of FSGS and help understand the mechanism of tubular and podocyte damage in FSGS (119). Yang and coworkers evaluated the concentration of urinary PLs extracted from 26 patients with chronic kidney disease (CKD) (including glomerulonephritis cases and tubulointerstitial injury cases), as well as 13 healthy controls (120). The results showed that some PC species [PC (16:0/16:0), PC (16:0/22:3), PC (16:0/18:1), and PC (16:0/18:2)] were significantly higher in the patient group than in the healthy control group.
Using the unilateral ureteral obstruction (UUO) rat model, Yoshioka and coworkers compared the urinary PL profiles before and after UUO for two weeks. They reported that PE and PG with monoenoic or dienoic fatty acyl chains [PE (34:1), PE (34:2), PE (32:1), PE (36:2), PG (34:1), PG(32:1), PG (34:2), and PG (36:2)] were significantly higher in the UOO group than in the control cases, and thus could be used as potential biomarkers for kidney injury caused by UOO (121). Regarding neurological disorders, a previous study (122) focused on urinary bis (monoacylglycerol) phosphate (BMP, a special LPA species) levels in the leucine-rich repeat kinase 2 mutation carrier (LRRK2 mutation, the most common genetic cause of Parkinson’s disease), revealing that four urinary BMP isoforms were significantly higher in LRRK2 mutation carriers than in non-carriers, correlating with worse cognitive status.
Future aspects
Despite dramatic advancements in techniques (especially MS) for the identification and quantification of urinary PLs and LPLs, allowing the detection of numerous urinary PL and LPL species, there remain several challenges for the reliable detection of PLs at markedly low concentrations, which have great potential as disease-specific biomarkers. This relies on continued improvements in sample preparation methods (especially for urinary exosome study), as well as detection methods. Furthermore, the poor inter- and intra-laboratory reproducibility of urinary PL and LPL quantification and biomarker validation should be closely considered owing to diverse methodologies and limited cohort sizes in previous studies. Furthermore, bioinformatics or analytical strategies for combining urinary lipidomic profiling with other omics (e.g., proteomics) are required to thoroughly understand the metabolic pathways involved in disease development.
Conclusions
PLs have diverse biological functions in cells and play important roles in disease development, thus presenting immense potential for use as biomarker candidates. Urine is a versatile clinical sample allowing convenient and noninvasive collection, and is considered an ideal source of novel PL biomarkers. Although present at extremely low concentrations, the detection of urinary PLs and LPLs is now feasible owing to the rapid development of analytical techniques, revealing attractive clinical prospects in the screening of biomarkers. In previous studies, several urinary PLs or urinary exosome PLs have been determined as biomarker candidates. With further method development for analyte detection and data processing, urinary PL applications will provide a promising future for disease biomarker screening in clinical practice.
Acknowledgments
The authors would like to thank the laboratory members of the Department of Urology at the Graduate School of Medicine at Kyoto University for their intellectual support. They also thank Ms. Junko Hirao, Yukari Maruhashi, and Eriko Komaki for their excellent technical assistance and Ms. Haruka Nishihara and Shiho Kayama for clinical research coordination. The Editage (editage.com) language editing service is appreciated for language editing. XL is supported by the China Scholarship Council for a PhD study.
Funding: This work was supported by Grants-in-Aid for Scientific Research (15K10587 and 18H02936) from the Japan Society for the Promotion of Science.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Kazutoshi Fujita) for the series “Urinary Biomarkers of Urothelial Malignancies” published in Translational Andrology and Urology. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1263
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1263). The series “Urinary Biomarkers of Urothelial Malignancies” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ryan D, Robards K, Prenzler PD, et al. Recent and potential developments in the analysis of urine: a review. Anal Chim Acta 2011;684:8-20. [Crossref] [PubMed]
- Khamis MM, Adamko DJ, El-Aneed A. Mass spectrometric based approaches in urine metabolomics and biomarker discovery. Mass Spectrom Rev 2017;36:115-34. [Crossref] [PubMed]
- Spener F, Lagarde M, Géloên A, et al. What is lipidomics? Eur J Lipid Sci Technol 2003;105:481-2. [Crossref]
- Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res 2003;44:1071-9. [Crossref] [PubMed]
- Dowhan W. Understanding phospholipid function: Why are there so many lipids? J Biol Chem 2017;292:10755-66. [Crossref] [PubMed]
- Bandu R, Mok HJ, Kim KP. Phospholipids as cancer biomarkers: Mass spectrometry-based analysis. Mass Spectrom Rev 2018;37:107-38. [Crossref] [PubMed]
- Rodgers AL, Jappie-Mahomed D, van Jaarsveld PJ. Testing the dogma that total phospholipid fatty acid composition of blood plays a role in kidney stone pathogenesis, using a high-low risk human model: results from a pilot study. Urolithiasis 2019;47:255-61. [Crossref] [PubMed]
- Küllenberg D, Taylor LA, Schneider M, et al. Health effects of dietary phospholipids. Lipids Health Dis 2012;11:3. [Crossref] [PubMed]
- Schisterman EF, Mumford SL, Chen Z, et al. Lipid concentrations and semen quality: the LIFE study. Andrology 2014;2:408-15. [Crossref] [PubMed]
- Folch J, Lees M, Stanley GS. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226:497-509. [Crossref] [PubMed]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 1959;37:911-7. [Crossref] [PubMed]
- Löfgren L, Forsberg GB, Stahlman M. The BUME method: a new rapid and simple chloroform-free method for total lipid extraction of animal tissue. Sci Rep 2016;6:27688. [Crossref] [PubMed]
- Löfgren L, Stahlman M, Forsberg GB, et al. The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J Lipid Res 2012;53:1690-700. [Crossref] [PubMed]
- Matyash V, Liebisch G, Kurzchalia TV, et al. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 2008;49:1137-46. [Crossref] [PubMed]
- Hara A, Radin NS. Lipid extraction of tissues with a low-toxicity solvent. 1978.
- Li X, Nakayama K, Goto T, et al. Comparative evaluation of the extraction and analysis of urinary phospholipids and lysophospholipids using MALDI-TOF/MS. Chem Phys Lipids 2019;223:104787 [Crossref] [PubMed]
- Tipthara P, Thongboonkerd V. Differential human urinary lipid profiles using various lipid-extraction protocols: MALDI-TOF and LIFT-TOF/TOF analyses. Sci Rep 2016;6:33756. [Crossref] [PubMed]
- Zhang X, Choi FF, Zhou Y, et al. Metabolite profiling of plasma and urine from rats with TNBS-induced acute colitis using UPLC-ESI-QTOF-MS-based metabonomics--a pilot study. FEBS J 2012;279:2322-38. [Crossref] [PubMed]
- Kim H, Ahn E, Moon MH. Profiling of human urinary phospholipids by nanoflow liquid chromatography/tandem mass spectrometry. Analyst 2008;133:1656-63. [Crossref] [PubMed]
- Schiller J, Muller M, Fuchs B, et al. 31P NMR spectroscopy of phospholipids: From micelles to membranes. Curr Anal Chem 2007;3:283-301. [Crossref]
- Her C, Filoti DI, McLean MA, et al. The charge properties of phospholipid nanodiscs. Biophys J 2016;111:989-98. [Crossref] [PubMed]
- Gunstone FD. Phospholipid technology and applications. Amsterdam: Elsevier, 2008.
- Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science 1972;175:720-31. [Crossref] [PubMed]
- Cai T, Yang F. Phospholipid and phospholipidomics in health and diseases. Singapore: Springer, 2018:177-202.
- Li Z, Agellon LB, Allen TM, et al. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab 2006;3:321-31. [Crossref] [PubMed]
- Botan A, Favela-Rosales F, Fuchs PF, et al. Toward Atomistic Resolution Structure of Phosphatidylcholine Headgroup and Glycerol Backbone at Different Ambient Conditions. J Phys Chem B 2015;119:15075-88. [Crossref] [PubMed]
- Banfield MJ, Barker JJ, Perry AC, et al. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 1998;6:1245-54. [Crossref] [PubMed]
- Udell CM, Rajakulendran T, Sicheri F, et al. Mechanistic principles of RAF kinase signaling. Cell Mol Life Sci 2011;68:553-65. [Crossref] [PubMed]
- Calzada E, Onguka O, Claypool SM. Phosphatidylethanolamine Metabolism in Health and Disease. Int Rev Cell Mol Biol 2016;321:29-88. [Crossref] [PubMed]
- Kooijman EE, Chupin V, de Kruijff B, et al. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 2003;4:162-74. [Crossref] [PubMed]
- Wang X, Devaiah SP, Zhang W, et al. Signaling functions of phosphatidic acid. Prog Lipid Res 2006;45:250-78. [Crossref] [PubMed]
- Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta 2006;1761:913-26. [Crossref] [PubMed]
- Kooijman EE, Tieleman DP, Testerink C, et al. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J Biol Chem 2007;282:11356-64. [Crossref] [PubMed]
- Zhao C, Du G, Skowronek K, et al. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat Cell Biol 2007;9:706-12. [Crossref] [PubMed]
- Rizzo MA, Shome K, Watkins SC, et al. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J Biol Chem 2000;275:23911-8. [Crossref] [PubMed]
- Foster DA, Salloum D, Menon D, et al. Phospholipase D and the maintenance of phosphatidic acid levels for regulation of mammalian target of rapamycin (mTOR). J Biol Chem 2014;289:22583-8. [Crossref] [PubMed]
- Sergeant S, Waite KA, Heravi J, et al. Phosphatidic acid regulates tyrosine phosphorylating activity in human neutrophils: enhancement of Fgr activity. J Biol Chem 2001;276:4737-46. [Crossref] [PubMed]
- Delon C, Manifava M, Wood E, et al. Sphingosine kinase 1 is an intracellular effector of phosphatidic acid. J Biol Chem 2004;279:44763-74. [Crossref] [PubMed]
- Fang Y, Vilella-Bach M, Bachmann R, et al. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 2001;294:1942-5. [Crossref] [PubMed]
- Han H, Qi R, Zhou JJ, et al. Regulation of the Hippo Pathway by Phosphatidic Acid-Mediated Lipid-Protein Interaction. Mol Cell 2018;72:328-40 e8.
- Han S, Huh J, Kim W, et al. Phospholipase D activates HIF-1-VEGF pathway via phosphatidic acid. Exp Mol Med 2014;46:e126 [Crossref] [PubMed]
- Jang JH, Lee CS, Hwang D, et al. Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog Lipid Res 2012;51:71-81. [Crossref] [PubMed]
- Reue K, Wang H. Mammalian lipin phosphatidic acid phosphatases in lipid synthesis and beyond: metabolic and inflammatory disorders. J Lipid Res 2019;60:728-33. [Crossref] [PubMed]
- Meana C, Garcia-Rostan G, Pena L, et al. The phosphatidic acid phosphatase lipin-1 facilitates inflammation-driven colon carcinogenesis. JCI Insight 2018;3:e97506 [Crossref] [PubMed]
- Huang H, Gao Q, Peng X, et al. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 2011;20:376-87. [Crossref] [PubMed]
- Kay JG, Grinstein S. Phosphatidylserine-mediated cellular signaling. Singapore: Springer, 2013:177-93.
- Birge RB, Boeltz S, Kumar S, et al. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ 2016;23:962-78. [Crossref] [PubMed]
- Hirai H, Natori S, Sekimizu K. Reversal by phosphatidylglycerol and cardiolipin of inhibition of transcription and replication by histones in vitro. Arch Biochem Biophys 1992;298:458-63. [Crossref] [PubMed]
- Zhong Q, Li G, Gvozdenovic-Jeremic J, et al. Up-regulation of the cell integrity pathway in saccharomyces cerevisiae suppresses temperature sensitivity of the pgs1Delta mutant. J Biol Chem 2007;282:15946-53. [Crossref] [PubMed]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2002;2:489-501. [Crossref] [PubMed]
- Tsui MM, York JD. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv Enzyme Regul 2010;50:324-37. [Crossref] [PubMed]
- Carnero A, Blanco-Aparicio C, Renner O, et al. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets 2008;8:187-98. [Crossref] [PubMed]
- Ma Q, Zhu C, Zhang W, et al. Mitochondrial PIP3-binding protein FUNDC2 supports platelet survival via AKT signaling pathway. Cell Death Differ 2019;26:321-31. [Crossref] [PubMed]
- Ratcliffe CDH, Siddiqui N, Coelho PP, et al. HGF-induced migration depends on the PI(3,4,5)P3-binding microexon-spliced variant of the Arf6 exchange factor cytohesin-1. J Cell Biol 2019;218:285-98. [Crossref] [PubMed]
- Takenawa T, Itoh T. Phosphoinositides, key molecules for regulation of actin cytoskeletal organization and membrane traffic from the plasma membrane. Biochim Biophys Acta 2001;1533:190-206. [Crossref] [PubMed]
- Bilanges B, Posor Y, Vanhaesebroeck B. PI3K isoforms in cell signalling and vesicle trafficking. Nat Rev Mol Cell Biol 2019;20:515-34. [Crossref] [PubMed]
- Schink KO, Tan KW, Stenmark H. Phosphoinositides in Control of Membrane Dynamics. Annu Rev Cell Dev Biol 2016;32:143-71. [Crossref] [PubMed]
- Phan TK, Williams SA, Bindra GK, et al. Phosphoinositides: multipurpose cellular lipids with emerging roles in cell death. Cell Death Differ 2019;26:781-93. [Crossref] [PubMed]
- Hollie NI, Cash JG, Matlib MA, et al. Micromolar changes in lysophosphatidylcholine concentration cause minor effects on mitochondrial permeability but major alterations in function. Biochim Biophys Acta 2014;1841:888-95. [Crossref] [PubMed]
- Law SH, Chan ML, Marathe GK, et al. An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int J Mol Sci 2019;20:1149. [Crossref] [PubMed]
- Tan M, Hao F, Xu X, et al. Lysophosphatidylcholine activates a novel PKD2-mediated signaling pathway that controls monocyte migration. Arterioscler Thromb Vasc Biol 2009;29:1376-82. [Crossref] [PubMed]
- Yang LV, Radu CG, Wang L, et al. Gi-independent macrophage chemotaxis to lysophosphatidylcholine via the immunoregulatory GPCR G2A. Blood 2005;105:1127-34. [Crossref] [PubMed]
- Nishina A, Kimura H, Sekiguchi A, et al. Lysophosphatidylethanolamine in Grifola frondosa as a neurotrophic activator via activation of MAPK. J Lipid Res 2006;47:1434-43. [Crossref] [PubMed]
- Park KS, Lee HY, Lee SY, et al. Lysophosphatidylethanolamine stimulates chemotactic migration and cellular invasion in SK-OV3 human ovarian cancer cells: involvement of pertussis toxin-sensitive G-protein coupled receptor. FEBS Lett 2007;581:4411-6. [Crossref] [PubMed]
- Park SJ, Lee KP, Kang S, et al. Lysophosphatidylethanolamine utilizes LPA(1) and CD97 in MDA-MB-231 breast cancer cells. Cell Signal 2013;25:2147-54. [Crossref] [PubMed]
- Ye X. Lysophospholipid signaling in the function and pathology of the reproductive system. Hum Reprod Update 2008;14:519-36. [Crossref] [PubMed]
- Houben AJ, Moolenaar WH. Autotaxin and LPA receptor signaling in cancer. Cancer Metastasis Rev 2011;30:557-65. [Crossref] [PubMed]
- Lin ME, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat 2010;91:130-8. [Crossref] [PubMed]
- Yatomi Y, Kurano M, Ikeda H, et al. Lysophospholipids in laboratory medicine. Proc Jpn Acad Ser B Phys Biol Sci 2018;94:373-89. [Crossref] [PubMed]
- Li YF, Li RS, Samuel SB, et al. Lysophospholipids and their G protein-coupled receptors in atherosclerosis. Front Biosci (Landmark Ed) 2016;21:70-88. [Crossref]
- Nishikawa M, Kurano M, Ikeda H, et al. Lysophosphatidylserine has bilateral effects on macrophages in the pathogenesis of atherosclerosis. J Atheroscler Thromb 2015;22:518-26. [Crossref] [PubMed]
- Park KS, Kim MK, Im DS, et al. Effect of lysophosphatidylglycerol on several signaling molecules in OVCAR-3 human ovarian cancer cells: involvement of pertussis toxin-sensitive G-protein coupled receptor. Biochem Pharmacol 2007;73:675-81. [Crossref] [PubMed]
- Shim JW, Jo SH, Kim SD, et al. Lysophosphatidylglycerol inhibits formyl peptide receptorlike-1-stimulated chemotactic migration and IL-1beta production from human phagocytes. Exp Mol Med 2009;41:584-91. [Crossref] [PubMed]
- Metz SA. Lysophosphatidylinositol, but not lysophosphatidic acid, stimulates insulin release: a possible role for phospholipase A2 but not de novo synthesis of lysophospholipid in pancreatic islet function. Biochem Biophys Res Commun 1986;138:720-7. [Crossref] [PubMed]
- Metz SA. Mobilization of cellular Ca2+ by lysophospholipids in rat islets of Langerhans. Biochim Biophys Acta 1988;968:239-52. [Crossref] [PubMed]
- Falasca M, Corda D. Elevated levels and mitogenic activity of lysophosphatidylinositol in k-ras-transformed epithelial cells. Eur J Biochem 1994;221:383-9. [Crossref] [PubMed]
- Falasca M, Iurisci C, Carvelli A, et al. Release of the mitogen lysophosphatidylinositol from H-Ras-transformed fibroblasts; a possible mechanism of autocrine control of cell proliferation. Oncogene 1998;16:2357-65. [Crossref] [PubMed]
- Henstridge CM, Balenga NA, Kargl J, et al. Minireview: recent developments in the physiology and pathology of the lysophosphatidylinositol-sensitive receptor GPR55. Mol Endocrinol 2011;25:1835-48. [Crossref] [PubMed]
- Arifin SA, Falasca M. Lysophosphatidylinositol Signalling and Metabolic Diseases. Metabolites 2016;6:6. [Crossref] [PubMed]
- Rockenfeller P, Koska M, Pietrocola F, et al. Phosphatidylethanolamine positively regulates autophagy and longevity. Cell Death Differ 2015;22:499-508. [Crossref] [PubMed]
- Furse S. Is phosphatidylglycerol essential for terrestrial life? J Chem Biol 2016;10:1-9. [Crossref] [PubMed]
- Welte MA, Gould AP. Lipid droplet functions beyond energy storage. Biochim Biophys Acta Mol Cell Biol Lipids 2017;1862:1260-72. [Crossref] [PubMed]
- Walther TC, Chung J, Farese RV Jr. Lipid Droplet Biogenesis. Annu Rev Cell Dev Biol 2017;33:491-510. [Crossref] [PubMed]
- Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 2019;20:137-55. [Crossref] [PubMed]
- Choudhary V, Golani G, Joshi AS, et al. Architecture of Lipid Droplets in Endoplasmic Reticulum Is Determined by Phospholipid Intrinsic Curvature. Curr Biol 2018;28:915-26.e9. [Crossref] [PubMed]
- Peterson BL, Cummings BS. A review of chromatographic methods for the assessment of phospholipids in biological samples. Biomed Chromatogr 2006;20:227-43. [Crossref] [PubMed]
- Fuchs B, Suss R, Teuber K, et al. Lipid analysis by thin-layer chromatography--a review of the current state. J Chromatogr A 2011;1218:2754-74. [Crossref] [PubMed]
- Schiller J, Arnold K. Application of high resolution 31P NMR spectroscopy to the characterization of the phospholipid composition of tissues and body fluids - a methodological review. Med Sci Monit 2002;8:MT205-22. [PubMed]
- Araseki M, Yamamoto K, Miyashita K. Oxidative stability of polyunsaturated fatty acid in phosphatidylcholine liposomes. Biosci Biotechnol Biochem 2002;66:2573-7. [Crossref] [PubMed]
- Min HK, Lim S, Chung BC, et al. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal Bioanal Chem 2011;399:823-30. [Crossref] [PubMed]
- Rockwell HE, Gao F, Chen EY, et al. Dynamic Assessment of Functional Lipidomic Analysis in Human Urine. Lipids 2016;51:875-86. [Crossref] [PubMed]
- Skotland T, Ekroos K, Kauhanen D, et al. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur J Cancer 2017;70:122-32. [Crossref] [PubMed]
- Byeon SK, Lee JY, Lee JS, et al. Lipidomic profiling of plasma and urine from patients with Gaucher disease during enzyme replacement therapy by nanoflow liquid chromatography-tandem mass spectrometry. J Chromatogr A 2015;1381:132-9. [Crossref] [PubMed]
- Yang JS, Lee JC, Byeon SK, et al. Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal Chem 2017;89:2488-96. [Crossref] [PubMed]
- Mirzoyan K, Baiotto A, Dupuy A, et al. Increased urinary lysophosphatidic acid in mouse with subtotal nephrectomy: potential involvement in chronic kidney disease. J Physiol Biochem 2016;72:803-12. [Crossref] [PubMed]
- Boonla C, Youngjermchan P, Pumpaisanchai S, et al. Lithogenic activity and clinical relevance of lipids extracted from urines and stones of nephrolithiasis patients. Urol Res 2011;39:9-19. [Crossref] [PubMed]
- Fuchs B. Analysis of phospolipids and glycolipids by thin-layer chromatography-matrix-assisted laser desorption and ionization mass spectrometry. J Chromatogr A 2012;1259:62-73. [Crossref] [PubMed]
- Clos-Garcia M, Loizaga-Iriarte A, Zuniga-Garcia P, et al. Metabolic alterations in urine extracellular vesicles are associated to prostate cancer pathogenesis and progression. J Extracell Vesicles 2018;7:1470442 [Crossref] [PubMed]
- Bang DY, Byeon SK, Moon MH. Rapid and simple extraction of lipids from blood plasma and urine for liquid chromatography-tandem mass spectrometry. J Chromatogr A 2014;1331:19-26. [Crossref] [PubMed]
- Pearce JM, Komoroski RA. Resolution of phospholipid molecular species by 31P NMR. Magn Reson Med 1993;29:724-31. [Crossref] [PubMed]
- Komoroski RA, Pearce JM, Griffin WS, et al. Phospholipid abnormalities in postmortem schizophrenic brains detected by 31P nuclear magnetic resonance spectroscopy: a preliminary study. Psychiatry Res 2001;106:171-80. [Crossref] [PubMed]
- Schröter J, Popkova Y, Suss R, et al. Combined Use of MALDI-TOF Mass Spectrometry and (31)P NMR Spectroscopy for Analysis of Phospholipids. Methods Mol Biol 2017;1609:107-22. [Crossref] [PubMed]
- Han X. Lipidomics: Comprehensive mass spectrometry of lipids. Hoboken, New Jersey: John Wiley & Sons, 2016.
- Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev 2005;24:367-412. [Crossref] [PubMed]
- Hsu FF. Mass spectrometry-based shotgun lipidomics - a critical review from the technical point of view. Anal Bioanal Chem 2018;410:6387-409. [Crossref] [PubMed]
- Fuchs B, Suss R, Schiller J. An update of MALDI-TOF mass spectrometry in lipid research. Prog Lipid Res 2010;49:450-75. [Crossref] [PubMed]
- Sun G, Yang K, Zhao Z, et al. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte− matrix interactions. Anal Chem 2008;80:7576-85. [Crossref] [PubMed]
- Bresler K, Pyttel S, Paasch U, et al. Parameters affecting the accuracy of the MALDI-TOF MS determination of the phosphatidylcholine/lysophosphatidylcholine (PC/LPC) ratio as potential marker of spermatozoa quality. Chem Phys Lipids 2011;164:696-702. [Crossref] [PubMed]
- Wang J, Wang C, Han X. Enhanced coverage of lipid analysis and imaging by matrix-assisted laser desorption/ionization mass spectrometry via a strategy with an optimized mixture of matrices. Anal Chim Acta 2018;1000:155-62. [Crossref] [PubMed]
- Leopold J, Popkova Y, Engel KM, et al. Recent Developments of Useful MALDI Matrices for the Mass Spectrometric Characterization of Lipids. Biomolecules 2018;8:173. [Crossref] [PubMed]
- Fuchs B, Schiller J, Suss R, et al. A direct and simple method of coupling matrix-assisted laser desorption and ionization time-of-flight mass spectrometry (MALDI-TOF MS) to thin-layer chromatography (TLC) for the analysis of phospholipids from egg yolk. Anal Bioanal Chem 2007;389:827-34. [Crossref] [PubMed]
- Teuber K, Riemer T, Schiller J. Thin-layer chromatography combined with MALDI-TOF-MS and 31P-NMR to study possible selective bindings of phospholipids to silica gel. Anal Bioanal Chem 2010;398:2833-42. [Crossref] [PubMed]
- Fuchs B. Analysis of phospolipids and glycolipids by thin-layer chromatography-matrix-assisted laser desorption and ionization mass spectrometry. J Chromatogr A 2012;1259:62-73. [Crossref] [PubMed]
- Del Boccio P, Raimondo F, Pieragostino D, et al. A hyphenated microLC-Q-TOF-MS platform for exosomal lipidomics investigations: application to RCC urinary exosomes. Electrophoresis 2012;33:689-96. [Crossref] [PubMed]
- Kim H, Min HK, Kong G, et al. Quantitative analysis of phosphatidylcholines and phosphatidylethanolamines in urine of patients with breast cancer by nanoflow liquid chromatography/tandem mass spectrometry. Anal Bioanal Chem 2009;393:1649-56. [Crossref] [PubMed]
- Min HK, Kong G, Moon MH. Quantitative analysis of urinary phospholipids found in patients with breast cancer by nanoflow liquid chromatography-tandem mass spectrometry: II. Negative ion mode analysis of four phospholipid classes. Anal Bioanal Chem 2010;396:1273-80. [Crossref] [PubMed]
- Cala M, Aldana J, Sanchez J, et al. Urinary metabolite and lipid alterations in Colombian Hispanic women with breast cancer: A pilot study. J Pharm Biomed Anal 2018;152:234-41. [Crossref] [PubMed]
- Saulnier-Blache JS, Feigerlova E, Halimi JM, et al. Urinary lysophopholipids are increased in diabetic patients with nephropathy. J Diabetes Complications 2017;31:1103-8. [Crossref] [PubMed]
- Erkan E, Zhao X, Setchell K, et al. Distinct urinary lipid profile in children with focal segmental glomerulosclerosis. Pediatr Nephrol 2016;31:581-8. [Crossref] [PubMed]
- Yang WL, Bai Q, Li DD, et al. Changes of urinary phospholipids in the chronic kidney disease patients. Biomarkers 2013;18:601-6. [Crossref] [PubMed]
- Yoshioka Y, Tsutsumi T, Adachi M, et al. Altered phospholipid profile in urine of rats with unilateral ureteral obstruction. Metabolomics 2009;5:429-33. [Crossref]
- Alcalay RN, Hsieh F, Tengstrand E, et al. Higher Urine bis(Monoacylglycerol)Phosphate Levels in LRRK2 G2019S Mutation Carriers: Implications for Therapeutic Development. Mov Disord 2020;35:134-41. [Crossref] [PubMed]
- Graessler J, Mehnert CS, Schulte KM, et al. Urinary Lipidomics: evidence for multiple sources and sexual dimorphism in healthy individuals. Pharmacogenomics J 2018;18:331-9. [Crossref] [PubMed]


