
Whole blood GRHL2 expression as a prognostic biomarker in metastatic hormone-sensitive and castration-resistant prostate cancer
Introduction
In recent years, the management of metastatic hormone-sensitive prostate cancer (mHSPC) has undergone major change. Early treatment intensification with docetaxel chemotherapy or potent androgen receptor pathway inhibitors (ARPI) at the commencement of androgen deprivation therapy (ADT) has resulted in profound improvements in overall survival (OS) and quality of life, representing the new standard-of-care for this disease entity (1). As these potent systemic therapies move earlier into the prostate cancer disease course, and more treatment choices become available, it is imperative that prognostic and predictive biomarkers be identified to guide optimal treatment selection for both new and existing agents.
Biomarkers in metastatic prostate cancer have traditionally centred around clinicopathological factors to predict prostate tumour biology and response to systemic treatment. In mCRPC, these characteristics have included lactate dehydrogenase (LDH), prostate-specific antigen (PSA) level, alkaline phosphatase (ALP), albumin, Gleason score, Eastern Cooperative Oncology Group (ECOG) performance status, haemoglobin, presence of visceral disease and use of opioid analgesia (2-4). Similar clinical parameters have also been found to be prognostic in the mHSPC population (5,6). While these clinical parameters demonstrate reasonable concordance with clinical outcomes, they are insufficient to influence treatment selections. Furthermore, as sequencing of systemic therapy appears to influence the efficacy of subsequent lines of treatment (7), there is even greater imperative that methods beyond clinical variables be used to personalise metastatic prostate cancer treatment. These shortcoming of clinicopathological variables as decision aids have led many to propose molecular characterisation of prostate cancer as an alternative pathway towards identification of promising candidate biomarkers capable of being incorporated into clinical practice.
The conventional approach to molecular biomarker identification entails analysis of tumour tissue samples. However, the nature of metastatic prostate cancer is such that obtaining adequate tumour material for analysis remains technically challenging. Attention has instead shifted to the development of minimally invasive blood-based biomarkers. To date, the field has focused primarily on the analysis of circulating tumour cells (CTCs) (8-10) and circulating tumour DNA (11,12), supported by advances in high-throughput sequencing technologies and increasingly sophisticated bioinformatic analysis techniques. Characterisation of whole blood RNA expression is another approach for molecular profiling in prostate cancer patients (13-16). This technique offers the opportunity to investigate diverse sources of RNA within blood including extracellular vesicles, platelets and circulating cell-free RNA (17), with the added benefit of being amenable to workflow automation, an important consideration that facilitates implementation in diagnostic laboratories outside of academic and research institutions (18,19).
We have previously developed a whole blood quantitative reverse transcription polymerase chain reaction (RT-PCR) assay to detect a panel of eight circulating prostate cancer-associated gene transcripts in a group of mCRPC patients (20). Building upon this work, we hypothesised that expression of specific transcripts may correlate with clinical outcomes related to tumour progression in mHSPC, and these same transcripts would have prognostic utility in our mCRPC cohort. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1444).
Methods
Study population and sample collection
This cohort study included two distinct patient groups: mHSPC and mCRPC. Patients with mHSPC were prospectively enrolled from two separate Australian institutions: Monash Health (March 2018–June 2019) and St Vincent’s Hospital (November 2016–October 2018). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Human Research Ethics Committee at Monash Health (HREC 11571X) and St Vincent’s Hospital (SVH 12/231). Written informed consent was obtained from all individual participants. Patients were eligible if initial blood draw occurred prior to or within six weeks of first administration of ADT (i.e., gonadotropin-releasing hormone analogues and/or first-generation antiandrogens). To optimise study recruitment, no other additional inclusion or exclusion criteria were mandated for this cohort.
The mCRPC cohort was prospectively enrolled from three Australian institutions (Monash Health, Chris O’Brien Lifehouse and Eastern Health) between September 2016 and September 2018, and has been previously described (20). Key eligibility criteria include patients with biochemically or radiographically progressive mCRPC planned for commencement of new systemic therapy, either ARPIs (e.g., abiraterone, enzalutamide) or taxane chemotherapy (e.g., docetaxel, cabazitaxel). Blood samples were taken immediately prior to the commencement of systemic therapy. Compared to prior reports (20), the present study represents an expanded patient sample with an extended follow-up period (up to September 30, 2019).
Gene expression assay
The RT-PCR assay was performed as previously described (20). Briefly, peripheral blood (2.5 mL) was collected in a PAXgene blood RNA tube (Qiagen, Hilden, Germany) and stored at –80 °C until batch sample processing. RNA was extracted from thawed whole blood, before undergoing qualitative and quantitative assessment. High-quality RNA was then reverse transcribed and pre-amplified. Gene expression was assessed using pre-designed, publicly available TaqmanTM assays for the following target transcripts: FOLH1, FOXA1, GRHL2, HOXB13, KLK2, KLK3, NPY and TMPRSS2:ERG. Gene expression was defined as samples that reached cycle threshold within 35 cycles in at least two of three replicates per patient.
Data collection, outcome measures and follow-up
In both the mHSPC and mCRPC cohorts, the following data was collected at the time of PAXgene sample collection and stored in a protected clinical database: prior local and systemic treatment, racial background, Eastern Cooperative Oncology Group (ECOG) performance status, disease extent, complete blood count and serum biochemistry (including PSA level). In the mHSPC cohort, additional clinical characteristics related to disease volume and use of treatment intensification were also collected, both of which are known to have prognostic/predictive significance (21-23). Disease volume was categorised as per ECOG 3805 CHAARTED criteria (22), with high-volume disease defined by the presence of at least four bone metastases (with at least one beyond the axial skeleton), and/or the presence of visceral metastases. Treatment intensification was defined by concurrent administration of either docetaxel chemotherapy or AR targeted therapy with concurrent ADT therapy for mHSPC.
In the mHSPC cohort, the intermediate outcomes of seven-month undetectable PSA rate (defined as ≤0.2 ng/mL) and time to castration-resistance (TTCR) were selected based on their established prognostic utility (24,25). TTCR was defined by duration between commencement of ADT and development of PSA, symptomatic or radiographic progression. In the mCRPC cohort, the outcome measures investigated were as follows: (I) PSA50 response (PSA decline from baseline of ≥50%, confirmed ≥3 weeks later), (II) progression-free survival (PFS) (time from treatment commencement to first of confirmed PSA progression, clinical or radiographic progression, or death from prostate cancer), and (III) OS (time from treatment commencement until death from any cause).
Follow-up interval was not strictly enforced for study patients, but left to the clinical discretion of the site investigator. In general, patients were reviewed monthly during ARPI therapy, three-weekly during taxane chemotherapy and three-monthly if receiving ADT monotherapy. Serial PSA measurements for disease monitoring were performed at the same laboratory prior to each follow-up visit, and repeat radiographic imaging was at the discretion of the investigator.
Statistical analysis
The associations between gene panel transcripts and categorical outcome measures (seven-month undetectable PSA in mHSPC cohort; PSA50 response in mCRPC cohort) were assessed using chi-square statistics. Univariable Cox proportional hazards models were used to assess the association between gene transcript expression and time-to-event outcomes (TTCR in mHSPC cohort; PFS and OS in mCRPC cohort), with P values adjusted for multiple testing using the Benjamini-Hochberg correction where appropriate (26). Exploratory subgroup analysis based on treatment administered (taxane chemotherapy and ARPI) was also performed in the mCRPC cohort. Gene transcripts reaching statistical significance (defined as P<0.05) in univariable analyses were incorporated into multivariable models with established clinical prognostic variables. Multivariable analyses in the mHSPC cohort (n=43) were restricted to three covariates to prevent overfitting: disease volume (high vs. low), use of treatment intensification, and haemoglobin [< lower limit of normal (LLN) vs. ≥ LLN]. Multivariable analyses in the mCRPC cohort (n=119) included prior chemotherapy exposure (yes vs. no), prior ARPI exposure (yes vs. no), haemoglobin (< LLN vs. ≥ LLN), Eastern Cooperative Oncology Group (ECOG) performance status (≥2 vs. 0–1), and presence of visceral metastases. All time-to-event outcomes were censored at date of last patient contact if the event had not occurred. All statistics and survival curves were generated using R Studio, version 1.1.463 (RStudio: Integrated Development Environment for R, Boston, Massachusetts). Given the exploratory nature of the study, no formal sample size calculations were undertaken. Instead, the study adopted a pragmatic design with few eligibility criteria, aimed at maximising patient recruitment.
Results
Study cohorts
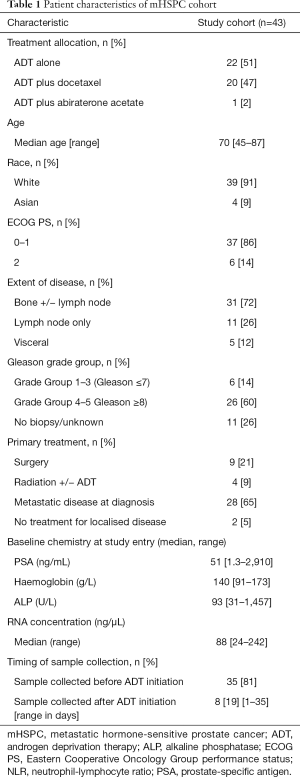
Peripheral whole blood was obtained from 43 mHSPC patients. The clinical characteristics of this cohort are shown in Table 1. A total of 65% (28/43) of patients had de novo metastases at mHSPC diagnosis, including 40% (17/43) with high-volume disease. Upfront treatment intensification was administered in 49% (21/43) of patients. The relative detection frequency for each gene transcript is presented in Figure S1. At the data cut-off date of September 30, 2019, median follow-up time in the mHSPC cohort was 13.5 months [interquartile range (IQR), 7.8–20.5 months (mo)] for living patients. In the mCRPC cohort, pre-treatment whole blood samples were obtained from 119 patients immediately prior to commencing AR pathway inhibitors (n=83) or taxane chemotherapy (n=36). The clinical characteristics of this cohort are presented in Table 2. At the data cut-off date of September 30, 2019, median follow-up time in the mCRPC cohort was 22.8 mo (IQR, 12.0–30.6 mo) for living patients.
Full table

Full table
Gene transcript expression and outcomes in mHSPC cohort
Of the 43 patients, 39 (91%) had available data to assess seven-month undetectable PSA as an outcome measure. Table S1 summarises the undetectable PSA rates according to gene panel transcripts. Patients with detectable GRHL2, HOXB13 or KLK3 transcripts in blood were less likely to achieve an undetectable PSA, but these associations were not statistically significant (GRHL2: 25% vs. 65%, P=0.24; HOXB13 33% vs. 66%, P=0.24; KLK3 38% vs. 65%, P=0.27; adjusted for multiple testing). There was no significant difference in use of treatment intensification based on GRHL2 status (GRHL2+ vs. GRHL2–: 50% vs. 49%, P=1.0), HOXB13 status (HOXB13+ vs. HOXB13–: 62% vs. 43%, P=0.3) or KLK3 status (KLK3+ vs. KLK3–: 57% vs. 45%, P=0.6), suggesting that use of docetaxel chemotherapy or ARPI were unlikely to have impacted these findings.
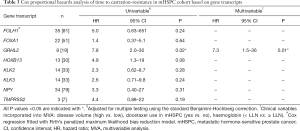
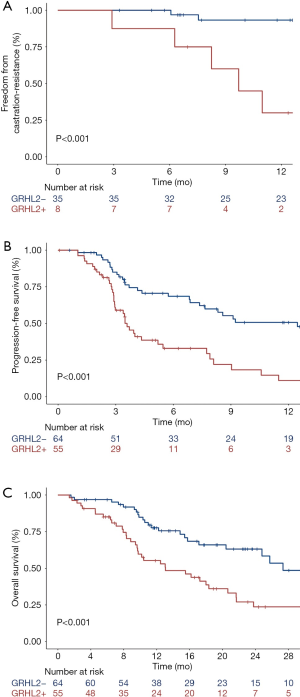
The median TTCR for the mHSPC cohort was not reached. Cox regression analysis of the association between gene transcripts and TTCR are shown in Table 3. In univariable analysis, circulating GRHL2 was strongly associated with shorter TTCR (median TTCR 9.7 mo vs. not reached; 12-month mCRPC progression rate 93% vs. 30%; P<0.001; Figure 1A). This remained significant in multivariable analysis when accounting for established clinicopathological prognostic markers of disease burden (Table S2) including disease volume, treatment intensification use and baseline haemoglobin (HR 7.3, 95% CI: 1.5–36, P=0.01). No other transcripts were significantly associated with TTCR.
Full table
GRHL2 expression and outcomes in mCRPC
Given the strong association of GRHL2 expression and outcomes observed in the mHSPC cohort, we tested clinical outcomes with GRHL2 expression in a separate cohort of mCRPC patients. In total, GRHL2-positive patients comprised 46% (55/119) of the cohort. Presence of the GRHL2 transcript predicted for significantly lower PSA50 response rates [positive/negative: 25/55 (46%) vs. 44/64 (69%), P=0.01]. Analysis by treatment administered demonstrated that these results were driven primarily by ARPI-treated patients [positive/negative: 16/37 (43%) vs. 35/46 (76%), P=0.003], with no effect apparent in taxane chemotherapy-treated patients [positive/negative: 9/18 (50%) vs. 9/18 (50%), P=1.0]. Figure S2 shows the waterfall plots of best PSA response according to GRHL2 status and treatment administered.
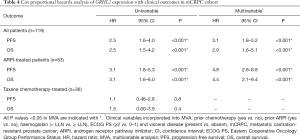
Median PFS and OS was 6.9 mo (IQR, 2.9–18.7 mo) and 20.6 mo (IQR, 9.8–34.5 mo), respectively. All but one patient in the mCRPC cohort died as a result of progressive prostate cancer. Patients with detectable circulating GRHL2 transcript experienced significantly shorter PFS and OS (Figure 1B,C). These associations remained significant in multivariable analysis when adjusting for other baseline prognostic clinicopathological factors (Table S3) including prior chemotherapy and ARPI exposure, ECOG performance status, visceral disease and baseline haemoglobin (Table 4); similar to PSA response, these correlations were primarily observed in the ARPI-treated subgroup.
Full table
Discussion
In this prospective study, using a previously optimised whole blood RT-PCR assay, our aim was to identify a circulating biomarker(s) linked to treatment outcome in mHSPC patients commencing ADT and a separate cohort of mCRPC patients receiving contemporary systemic therapy. We found that detectable expression of the AR coregulator GRHL2 in blood was a negative prognostic biomarker throughout the clinical spectrum of metastatic prostate cancer and was associated with rapid resistance to ADT, shorter response to systemic therapy and reduced OS. Notably, these associations were present even when accounting for baseline clinicopathological variables of prognostic significance.
The GRHL2 protein is part of the grainyhead-like transcription factor family and has been implicated in the pathogenesis of various cancers (27), though its functional relevance in prostate cancer has remained largely undefined until recently. Critical work by Paltoglou et al. supports dual oncogenic and protective roles of GRHL2, enhancing AR signalling through its function as an AR coregulator, whilst simultaneously downregulating intracellular signalling pathways responsible for epithelial-mesenchymal transition and cellular invasion (28). This delicate counterbalance between oncogenic and tumour suppressive phenotypes has also been observed in ovarian and breast cancer (29,30). The results here suggest that the balance in prostate cancer may be tipped towards GRHL2 as an oncogenic driver, though validation in larger independent cohorts is necessary before arriving at any definitive conclusions.
There is a relative paucity of data on the prognostic significance of circulating GRHL2 in prostate cancer. Danila et al. selected GRHL2 as a part of a five-gene whole blood RT-PCR assay, which also included the transcripts KLK2, KLK3, HOXB13 and FOXA1 (31). The authors found that presence of two or more transcripts was associated with shorter OS, and had superior prognostic value compared to CTC enumeration alone (31). However, the contribution of each individual transcript to the overall impact of this composite signature was not reported. Todenhofer et al. addressed the relative contribution of these transcripts in a cohort of mCRPC patients exclusively treated with abiraterone acetate (32). While there was a strong trend towards GRHL2 expression and shorter OS, it did not correlate with other clinical endpoints, perhaps reflecting the relatively small sample size of 37 patients. Importantly, both these studies were exclusively in mCRPC population groups. To date, no data exists for GRHL2 in earlier disease states.
This study explored the relevance of GRHL2 in an mHSPC cohort. Notably, GRHL2-positive patients experienced particularly poor outcomes, rapidly progressing from ADT commencement to mCRPC (median, 9.7 mo). We also observed poorer outcomes in mCRPC patients with detectable circulating GRHL2 mRNA expression. These associations were most apparent in patients receiving ARPI therapy, consistent with preclinical evidence demonstrating GRHL2’s role in enhancing transcriptional activity of both full length AR and constitutively active truncated AR variants (28), which are well-recognised resistance mechanisms to AR directed therapies (11,33).
Circulating GRHL2 expression may reflect a tumour’s increased reliance on the AR pathway as a primary method of growth signalling. Whilst theoretically this may render such tumours more susceptible to AR directed therapies, the presence of AR gain/amplification in plasma circulating tumour DNA and circulating tumour RNA (ctDNA/ctRNA) next-generation sequencing assays has consistently been shown to be a negative predictive biomarker for response to both abiraterone acetate and enzalutamide (11,33-37). Similarly, detectable GRHL2 mRNA in the mHSPC cohort could suggest greater dependency on AR-mediated proliferation that is incompletely suppressed with LHRH analogue monotherapy. Given the association between greater levels of testosterone suppression and improved clinical outcomes (38), it is feasible that GRHL2-positive mHSPC patients would benefit from more intense androgen suppression compared to GRHL2-negative patients. Multiple phase 3 trials have demonstrated that treatment intensification in the form of ADT combined with AR-directed therapies significantly improves OS in patients with mHSPC (39-44). Whether mHSPC patients with circulating GRHL2 expression may preferentially benefit from these novel intensified AR inhibition approaches is an intriguing proposition worthy of greater exploration. Regardless, the established ability of GRHL2 to drive prostate cancer growth and disease progression in the castration-resistant environment, together with the data described here, justify further research into GRHL2 as a circulating biomarker candidate in metastatic prostate cancer.
Our study has several limitations. Small sample size especially in the mHSPC cohort, together with heterogeneity of treatment administered may impact the ability to detect meaningful associations and the overall generalisability of findings. Follow-up periods were relatively short, most notably in the mHSPC cohort, limiting the analysis of longer-term, more robust clinical outcomes such as OS. Future studies in independent cohorts are planned to ensure these results are reproducible and not just reflective of cohort-specific findings.
Conclusions
We demonstrated the utility of circulating GRHL2 as a negative prognostic biomarker across the clinical spectrum of metastatic prostate cancer. GRHL2 was associated with rapid resistance to ADT, shorter duration of response to novel AR directed therapies, more rapid tumour progression and poorer OS. These data support further investigation of GRHL2 as a candidate prognostic biomarker in prostate cancer and expansion of efforts to better understand the role of GRHL2 in mediating resistance to prostate cancer systemic therapy.
Acknowledgments
We thank the patients and families for their participation in this study. We also thank Ms. Sophie Beck for providing statistical support during the manuscript revision process.
Funding: The following authors formally acknowledge their respective sources of funding support: Edmond M. Kwan: NHMRC Postgraduate Scholarship, Monash University Postgraduate Publications Award; Heidi Fettke: Australian Government Research Training Program (RTP) Scholarship, Monash University Postgraduate Publications Award; Megan Crumbaker: Australian Government RTP Scholarship; Sarah Q. To: NHMRC CJ Martin Early Career Fellowship (APP1070112); Kate Mahon: Movember/Prostate Cancer Foundation of Australia Clinical Scientist Fellow; Ian D. Davis: NHMRC Practitioner Fellowship (APP1102604); Carmel Pezaro: Cancer Council Victoria Early Career Seed Grant; Lisa G. Horvath: Astellas Investigator-Initiated Grant, Cancer Institute NSW Translational Program Grant; Arun A. Azad: NHMRC Project Grant (GNT1098647), Victorian Cancer Agency Clinical Research Fellowship (CRF14009), Astellas Investigator-Initiated Grant.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1444
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1444
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1444). TT serves as an unpaid editorial board member of Translational Andrology and Urology from Aug 2019 to Jul 2021. EMK reports receiving honoraria from Janssen and Ipsen; travel & accommodation from Astellas Pharma, Pfizer and Ipsen; and institutional research funding from Astellas Pharma, AstraZeneca, Bristol Myers Squibb, Pfizer, and Merck Serono. AAA reports receiving compensation as a Consultant from Astellas Pharma, Janssen, and Novartis; speakers bureau for Astellas, Janssen, Novartis, Amgen, Ipsen, Bristol Myers Squibb, Merck Serono and Bayer; honoraria from Astellas, Novartis, Sanofi, AstraZeneca, Tolmar, Telix; Merck Serono; Janssen, Bristol Myers Squibb, Ipsen, Bayer, Pfizer, Amgen, Noxopharm, and Merck Sharpe Dome; research funding from Astellas (investigator), Merck Serono (investigator), Astra Zeneca (investigator), Bristol Myers Squibb (institutional), Astra Zeneca (institutional), Aptevo Therapeutics (institutional), Glaxo Smith Kline (institutional), Pfizer (institutional), MedImmune (institutional), Astellas (institutional), SYNthorx (institutional), Bionomics (institutional), Sanofi Aventis (institutional), Novartis (institutional), and Ipsen (institutional); travel and accommodation from Astellas, Merck Serono, Amgen, Novartis, Janssen, Tolmar, Pfizer;and is on the Scientific Advisory Board for Astellas, Novartis, Sanofi, AstraZeneca, Tolmar, Pfizer, Telix; Merck Serono; Janssen, Bristol Myers Squibb, Ipsen, Bayer, Merck Sharpe Dome, Amgen and Noxopharm. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Human Research Ethics Committee at Monash Health (HREC 11571X) and St Vincent’s Hospital (SVH 12/231). Written informed consent was obtained from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kwan EM, Thangasamy IA, Teh J, et al. Navigating systemic therapy for metastatic castration-naive prostate cancer. World J Urol 2021;39:339-48. [Crossref] [PubMed]
- Halabi S, Small EJ, Kantoff PW, et al. Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J Clin Oncol 2003;21:1232-7. [Crossref] [PubMed]
- Smaletz O, Scher HI, Small EJ, et al. Nomogram for overall survival of patients with progressive metastatic prostate cancer after castration. J Clin Oncol 2002;20:3972-82. [Crossref] [PubMed]
- Halabi S, Lin CY, Kelly WK, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol 2014;32:671-7. [Crossref] [PubMed]
- Gravis G, Boher JM, Fizazi K, et al. Prognostic Factors for Survival in Noncastrate Metastatic Prostate Cancer: Validation of the Glass Model and Development of a Novel Simplified Prognostic Model. Eur Urol 2015;68:196-204. [Crossref] [PubMed]
- Glass TR, Tangen CM, Crawford ED, et al. Metastatic carcinoma of the prostate: identifying prognostic groups using recursive partitioning. J Urol 2003;169:164-9. [Crossref] [PubMed]
- Delanoy N, Hardy-Bessard AC, Efstathiou E, et al. Sequencing of Taxanes and New Androgen-targeted Therapies in Metastatic Castration-resistant Prostate Cancer: Results of the International Multicentre Retrospective CATS Database. Eur Urol Oncol 2018;1:467-75. [Crossref] [PubMed]
- Scher HI, Lu D, Schreiber NA, et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol 2016;2:1441-9. [Crossref] [PubMed]
- Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028-38. [Crossref] [PubMed]
- De Laere B, van Dam PJ, Whitington T, et al. Comprehensive Profiling of the Androgen Receptor in Liquid Biopsies from Castration-resistant Prostate Cancer Reveals Novel Intra-AR Structural Variation and Splice Variant Expression Patterns. Eur Urol 2017;72:192-200. [Crossref] [PubMed]
- Romanel A, Gasi Tandefelt D, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med 2015;7:312re10 [Crossref] [PubMed]
- Annala M, Vandekerkhove G, Khalaf D, et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov 2018;8:444-57. [Crossref] [PubMed]
- Ross RW, Galsky MD, Scher HI, et al. A whole-blood RNA transcript-based prognostic model in men with castration-resistant prostate cancer: a prospective study. Lancet Oncol 2012;13:1105-13. [Crossref] [PubMed]
- Olmos D, Brewer D, Clark J, et al. Prognostic value of blood mRNA expression signatures in castration-resistant prostate cancer: a prospective, two-stage study. Lancet Oncol 2012;13:1114-24. [Crossref] [PubMed]
- Liu X, Ledet E, Li D, et al. A Whole Blood Assay for AR-V7 and AR(v567es) in Patients with Prostate Cancer. J Urol 2016;196:1758-63. [Crossref] [PubMed]
- To SQ, Kwan EM, Fettke HC, et al. Expression of Androgen Receptor Splice Variant 7 or 9 in Whole Blood Does Not Predict Response to Androgen-Axis-targeting Agents in Metastatic Castration-resistant Prostate Cancer. Eur Urol 2018;73:818-21. [Crossref] [PubMed]
- Junqueira-Neto S, Batista IA, Costa JL, et al. Liquid Biopsy beyond Circulating Tumor Cells and Cell-Free DNA. Acta Cytol 2019;63:479-88. [Crossref] [PubMed]
- Ilie M, Hofman V, Long E, et al. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann Transl Med 2014;2:107. [PubMed]
- Duffy MJ, Sturgeon CM, Soletormos G, et al. Validation of new cancer biomarkers: a position statement from the European group on tumor markers. Clin Chem 2015;61:809-20. [Crossref] [PubMed]
- Kwan EM, Fettke H, Docanto MM, et al. Prognostic Utility of a Whole-blood Androgen Receptor-based Gene Signature in Metastatic Castration-resistant Prostate Cancer. Eur Urol Focus 2021;7:63-70. [Crossref] [PubMed]
- Gravis G, Boher JM, Chen YH, et al. Burden of Metastatic Castrate Naive Prostate Cancer Patients, to Identify Men More Likely to Benefit from Early Docetaxel: Further Analyses of CHAARTED and GETUG-AFU15 Studies. Eur Urol 2018;73:847-55. [Crossref] [PubMed]
- Sweeney CJ, Chen YH, Carducci M, et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. N Engl J Med 2015;373:737-46. [Crossref] [PubMed]
- James ND, Sydes MR, Clarke NW, et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet 2016;387:1163-77. [Crossref] [PubMed]
- Harshman LC, Chen YH, Liu G, et al. Seven-Month Prostate-Specific Antigen Is Prognostic in Metastatic Hormone-Sensitive Prostate Cancer Treated With Androgen Deprivation With or Without Docetaxel. J Clin Oncol 2018;36:376-82. [Crossref] [PubMed]
- Bournakis E, Efstathiou E, Varkaris A, et al. Time to castration resistance is an independent predictor of castration-resistant prostate cancer survival. Anticancer Res 2011;31:1475-82. [PubMed]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289-300. [Crossref]
- Reese RM, Harrison MM, Alarid ET. Grainyhead-like Protein 2: The Emerging Role in Hormone-Dependent Cancers and Epigenetics. Endocrinology 2019;160:1275-88. [Crossref] [PubMed]
- Paltoglou S, Das R, Townley SL, et al. Novel Androgen Receptor Coregulator GRHL2 Exerts Both Oncogenic and Antimetastatic Functions in Prostate Cancer. Cancer Res 2017;77:3417-30. [Crossref] [PubMed]
- Faddaoui A, Sheta R, Bachvarova M, et al. Suppression of the grainyhead transcription factor 2 gene (GRHL2) inhibits the proliferation, migration, invasion and mediates cell cycle arrest of ovarian cancer cells. Cell Cycle 2017;16:693-706. [Crossref] [PubMed]
- Werner S, Frey S, Riethdorf S, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem 2013;288:22993-3008. [Crossref] [PubMed]
- Danila DC, Anand A, Schultz N, et al. Analytic and clinical validation of a prostate cancer-enhanced messenger RNA detection assay in whole blood as a prognostic biomarker for survival. Eur Urol 2014;65:1191-7. [Crossref] [PubMed]
- Todenhöfer T, Azad A, Stewart C, et al. AR-V7 Transcripts in Whole Blood RNA of Patients with Metastatic Castration Resistant Prostate Cancer Correlate with Response to Abiraterone Acetate. J Urol 2017;197:135-42. [Crossref] [PubMed]
- Azad AA, Volik SV, Wyatt AW, et al. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin Cancer Res 2015;21:2315-24. [Crossref] [PubMed]
- Carreira S, Romanel A, Goodall J, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med 2014;6:254ra125 [Crossref] [PubMed]
- Conteduca V, Wetterskog D, Sharabiani MTA, et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann Oncol 2017;28:1508-16. [Crossref] [PubMed]
- Wyatt AW, Annala M, Aggarwal R, et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst 2017;109:djx118 [Crossref] [PubMed]
- Fettke H, Kwan EM, Docanto MM, et al. Combined Cell-free DNA and RNA Profiling of the Androgen Receptor: Clinical Utility of a Novel Multianalyte Liquid Biopsy Assay for Metastatic Prostate Cancer. Eur Urol 2020;78:173-80. [Crossref] [PubMed]
- Crawford ED, Heidenreich A, Lawrentschuk N, et al. Androgen-targeted therapy in men with prostate cancer: evolving practice and future considerations. Prostate Cancer Prostatic Dis 2019;22:24-38. [Crossref] [PubMed]
- Fizazi K, Tran N, Fein L, et al. Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med 2017;377:352-60. [Crossref] [PubMed]
- Fizazi K, Tran N, Fein L, et al. Abiraterone acetate plus prednisone in patients with newly diagnosed high-risk metastatic castration-sensitive prostate cancer (LATITUDE): final overall survival analysis of a randomised, double-blind, phase 3 trial. Lancet Oncol 2019;20:686-700. [Crossref] [PubMed]
- James ND, de Bono JS, Spears MR, et al. Abiraterone for Prostate Cancer Not Previously Treated with Hormone Therapy. N Engl J Med 2017;377:338-51. [Crossref] [PubMed]
- Davis ID, Martin AJ, Stockler MR, et al. Enzalutamide with Standard First-Line Therapy in Metastatic Prostate Cancer. N Engl J Med 2019;381:121-31. [Crossref] [PubMed]
- Chi KN, Agarwal N, Bjartell A, et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med 2019;381:13-24. [Crossref] [PubMed]
- Armstrong AJ, Szmulewitz RZ, Petrylak DP, et al. ARCHES: A Randomized, Phase III Study of Androgen Deprivation Therapy With Enzalutamide or Placebo in Men With Metastatic Hormone-Sensitive Prostate Cancer. J Clin Oncol 2019;37:2974-86. [Crossref] [PubMed]


