Comprehensive analysis of the lncRNA-miRNA-mRNA regulatory network for bladder cancer
Introduction
Each year, there are 74,000 newly diagnosed bladder cancer (BC) patients in the USA and 430,000 worldwide, making it the fourth and 11th most common cancer among males and females, respectively (1,2). BC is divided into non-muscle-invasive BC (NMIBC) and muscle-invasive BC (MIBC). However, despite BC being the leading cancer type among all human cancers, most of BC patients are not well managed. According to an analysis of 2012 Surveillance, Epidemiology, and End Results Program data, 4,790 BC patients with high-grade NMIBC were diagnosed from 1992 to 2002, but only 1 patient received the recommended treatment (3). Despite advances in BC therapeutic regimens (e.g., chemotherapy, surgical, and radiotherapy), only about 60% of MIBC patients have a 5-year survival. The underlying pathogenesis of BC may be responsible for this; however, it is not well understood (4).
Tumorigenesis is a complex process significantly related to various genetic mutations, epigenetic alterations and chromosomal translocations (5). Non-coding RNAs (ncRNAs) are coded by the genome, but most of ncRNAs cannot be translated to proteins (6). Despite not being translated into proteins, ncRNAs have an essential role in multiple biological processes (BPs) (7). Long ncRNAs (lncRNAs; >200 nt) may be the most critical regulator of gene expression, cell growth, cell differentiation, cell development, and chromatin dynamics (8). However, the biological functions of lncRNAs still be needed to be further classified (9). Thousands of lncRNAs have been found to have aberrant expression or mutations in multiple cancer types (10). Prensner et al. revealed that prostate cancer antigen 3, prostate cancer gene expression marker 1, and prostate cancer-associated ncRNA transcript 1 (PCAT-1) have been observed in prostate cancer (11). PCAT-1 promotes prostate cancer cell proliferation via the upregulation of c-Myc in the post-transcriptional phase (12). The aberrant expression of lncRNAs have also been observed in breast cancer, lung cancer, colorectal cancer, and BC (13-16). The role of lncRNAs may differ in various cancers; even the same lncRNAs could have different biological functions in different cancers. The overexpression of MEG3 could accelerate apoptosis and inhibit the proliferation of BC and lung cancer cells (17). However, the overexpression of linc-RoR has been found to promote epithelial-mesenchymal transition (EMT), drug resistance, and invasiveness of breast, pancreatic, and hepatocellular cancer cells (18). One of the significant molecular mechanisms of lncRNAs is competing endogenous RNAs (ceRNAs), which act as sponges of microRNA (miRNA). The aberrant expression of miRNAs has also found in many cancer types (19,20). Furthermore, they are significant regulators of tumorigenesis, progression, tumor suppressor, and drug resistance for various cancer types (21-23). Therefore, lncRNAs and miRNAs play an essential role in tumorigenesis, progression, and resistance. However, interactions between lncRNAs, miRNAs, and messenger RNAs (mRNA) still be needed to further investigated in BC.
In the present study, we investigated the potential biological functions and underlying pathological mechanisms of distinct lncRNAs, miRNAs, and mRNAs for BC using computational biology, further constructing a network to explore their relationship. The findings provide significant insight into the molecular mechanisms. Compared to Lyu et al. (24) and Wang et al. (25) studys, we not only investigated the Immune infiltration status between bladder cancer and normal tissue, we also explored the correlation expressed between the mRNA in BC ceRNA network and immune cells. Furthermore, we explored the immune cell types that significantly influence bladder cancer prognosis.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-21-81).
Methods
Data preparation and differentially expressed gene analysis
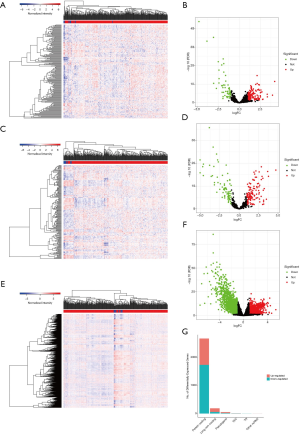
Raw data of BC were collected from The Cancer Genome Atlas (TCGA) including transcriptome profiling and clinical data. Limma package (version 3.44.0) in R software (https://www.r-project.org/) was used to analyze and identify differentially expressed RNAs (including DEmRNAs and DElncRNAs) and DEmiRNAs with thresholds of |logFC| >1.0 and false discovery rate (FDR<0.05). The package ggplot2 was used to construct the heatmap and volcano plot.
Construction of the ceRNA network
We constructed a ceRNA network base on the ceRNA hypothesis. Relevant miRNA target genes were obtained from StarBase, and only the targets which supported by experiments, including luciferase reporter assay, Western blot, Northern blot, or quantitative reverse transcription polymerase chain reaction, those we defined as the target genes. Only the DEmiRNAs were using to predict the target gene.
Biomarker screening and validation
The survival data of BC patients were extracted from TCGA. Subsequently, the RNAs identified in ceRNA network were selected for screening biomarkers. We used univariate Cox regression to screen prognostic RNAs (P<0.05), and prognostic RNAs with expression levels significantly relevant to patients’ overall survival (P<0.05) were selected as primitive biomarkers.
Cox risk regression establishment and validation
The raw data of lncRNAs, mRNAs, and miRNAs were transformed and normalized in a log2(cpm[x]+1) manner. Base on univariate Cox regression prognostic biomarkers, we performed multivariate Cox regression analysis combined with stepwise regression to establish a Cox risk model. We randomly selected half of the BC (n=404) samples to establish the risk model, another half of samples set and the total samples set were applied to validate the accuracy of the model. Finally, we identified the risk score as an independent prognostic factor from the clinical traits.
Protein-protein interaction analysis
DEmRNAs in the ceRNA network were used in the protein-protein interaction (PPI) network through the STRING (version 11.0) database (https://string-db.org/), with a confidence score >0.4.
Functional enrichment analysis
ClusterProfiler package in R was used for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology enrichment analyses, including BP, cellular component (CC), and molecular function (MF). The ggplot2 and nrichplot packages were used to visualize the enrichment results.
Immune infiltration analysis
To explore the infiltration of immune cells into the tumor microenvironment, we used CIBERSORT to estimate the abundance profile of immune cells in 414 tumor samples, we only selected the P value of CIBERSORT algorithm less than 0.05 for the infiltration analysis, which followed by only 175 tumor samples. Furthermore, we used the TIMER database to explore the relationship between the significant influence on prognosis in DEmRNAs and immune cell infiltration in BC, and investigated the prognosis of immune cells in BC.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Statistical analysis
The differential genes were identified via limma package in R software (https://www.r-project.org/). The threshold for screening significantly expressed genes was set as log fold change |logFC| >1 and false discovery rate FDR <0.05; The StarBase database was used to predict the miRNA target genes (http://starbase.sysu.edu.cn/), and the ceRNA network were visualization by Cytoscape software (vision 3.8.1). The Kaplan-Meier method was used to perform the survival curves. Enrichment analysis (GO and KEGG) were used the cluterProfiler and enrichplot package; The PPI network was constructed via the STRING website with a confidence score >0.4; The CIBERSORT package in R and TIMER database (https://cistrome.shinyapps.io/timer/) were used to estimate the tumor infiltrating immune cells. P value <0.05 was seem as the significant difference.
Results
Differential expression of lncRNAs, miRNAs, and mRNAs
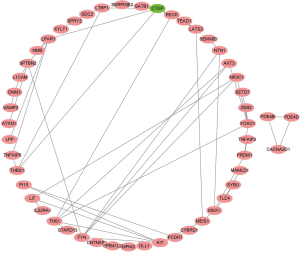
The differential expression analysis results showed that there were 59 downregulated and 129 upregulated lncRNAs, 121 upregulated and 79 downregulated miRNAs, and 1,726 downregulated and 935 upregulated mRNAs significantly expressed in the TGCA BC cohort, respectively (P<0.05) (Figure 1).
Construction of the ceRNA network
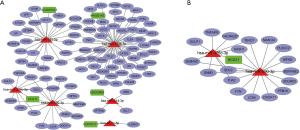
We further used the StarBase dataset to investigate the interaction of DElncRNAs, DEmiRNAs, and DEmRNAs. The results showed that 6 DEmiRNAs (has-mir-18a-5p, has-mir-347b-5p, has-mir-376c-3p, has-mir-590-3p, has-mir-214-3p, and has-let-7c-5p), 5 DElncRNAs (AC093010.3, MAGI2-AS3, HCG11, LINC00958, and ACO74117), and 97 DEmRNA were included in the ceRNA network (Figure 2A). One subnetwork showed that the potential interaction between lncRNA and miRNA and mRNA, such as lncRNA HCG11 (HLA complex group 11) may act as a sponge for has-miR-376c-3p to regulated mRNA Calcium Voltage-gated Channel Auxiliary Subunit Alpha2delta1 (CACNA2D1) (Figure 2B). The findings indicated that DElncRNAs regulated mRNA expression via interaction with miRNAs.
Cox model analysis
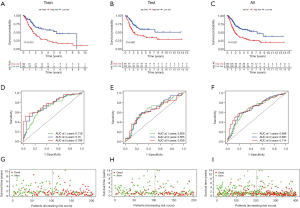
To explore the association DElncRNAs, DEmiRNAs, and DEmRNAs with the prognosis of BC, we used lncRNAs, miRNAs, and mRNAs in the ceRNA network to perform a Cox regression analysis combined with stepwise regression to establish a Cox risk model. There were 404 patients with complete clinical information be enrolled in the prognosis analyzed.
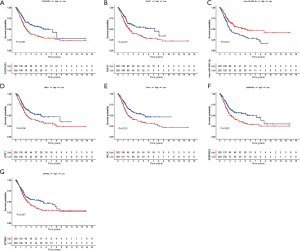
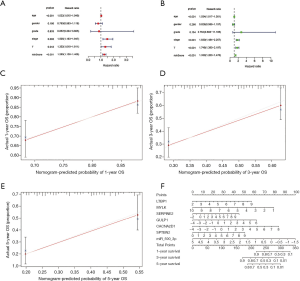
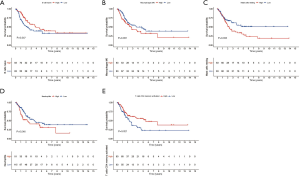
Training cohort showed that the high-risk patients had a shorter overall survival, the test cohort and all samples cohort also revealed that high-risk patients had a worse prognosis. (Figure 3A,B,C). These models predicted the correct rates of 1-year overall survival [area under the curve (AUC) for all samples cohort: 0.688, training cohort AUC: 0.719, test cohort AUC: 0.659], 3-year overall survival (AUC for all samples cohort: 0.697, training cohort AUC: 0.75, test cohort AUC: 0.655), and 5-year overall survival (AUC for all samples cohort: 0.714, training cohort AUC: 0.788, test cohort AUC: 0.639), respectively (Figure 3D,E,F). The results of survival status of 3 cohorts showed in Figure 3G,H,I. These results showed that our model can be a good predictor for BC prognosis. There were six DEmRNAs [CACNA2D1: P<0.001, domain containing engulfment adaptor1 (GULP1): P=0.001, latent transforming growth factor beta binding protein 1 (LTBP1): P=0.006, myosin light chain kinase (MYLK): P=0.001, serpin family E member 2 (SERPINE2): P=0.002, spectrin beta non-erythrocytic 2 (SPTBN2): P=0.047] and 1 DEmiRNA (hsa-miR-590-3p: P<0.001) with the significant influence on BC patients’ prognosis in our risk model (Figure 4). The overexpression of CACNA2D1, GULP1, LTBP1, MYLK, SERPINE2, and SPTBN2 led to poor BC prognosis, but the overexpression of hsa-miR-590-3p appeared to be a favorable biomarker. We used univariate and multivariate analyses to determine the association between risk scores and clinical features. The findings indicated that this risk model was an independent predictive factor for BC prognosis (Figure 5).
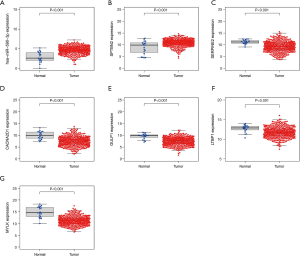
Expression levels of individual genes of risk model between tumor and normal tissue
Compared to normal samples, hsa-miR-590-3p and SPTBN2 (P<0.001) were significantly upregulated in tumor samples (Figure 6A,B). The findings indicated that SERPINE2 (P<0.001), CACNA2D1 (P<0.001), GULP1 (P<0.001), LTBP1 (P<0.001), and MYLK (P<0.001) were significantly overexpressed in normal samples than BC samples (Figure 6C,D,E,F,G). These results indicated that these genes may also be a diagnostics biomarker for BC patients.
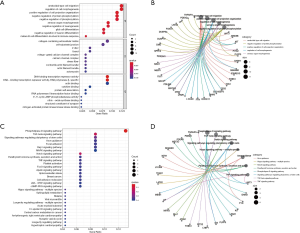
Functional enrichment analysis
We explored the potential biological functions of ceRNA. The findings indicated that the top 5 BPs of DElncRNAs, DEmiRNAs, and DEmRNAs were the negative regulation of protein phosphorylation, positive regulation of cell projection organization, regulation of cell morphogenesis, ameboidal-type cell migration, and negative regulation of phosphorylation. The top 5 CCs were voltage-gated calcium channel complex, I band, Z disc, cell-substrate junction, and collagen-containing extracellular matrix. The top 5 MFs were cytokine binding, actin binding, DNA-binding transcription repressor activity, RNA polymerase II-specific DNA-binding transcription repressor activity, and protein self-association (Figure 7A,B,C). The results of the KEGG pathway analysis indicated that the top 10 pathways of DEmRNAs were parathyroid hormone synthesis, secretion and action, the notch signaling pathway, the mitogen-activated protein kinase signaling pathway (MAPK), the Rap1 signaling pathway, focal adhesion, axon guidance, signaling pathways regulating the pluripotency of stem cells, the transforming growth factor-β (TGF-β) signaling pathway, the phospholipase D signaling pathway, and the tumor necrosis factor (TNF) signaling pathway (Figure 7C,D). The PPI network of the ceRNA network’s messenger RNA is shown in Figure 8.
Immune infiltration analysis
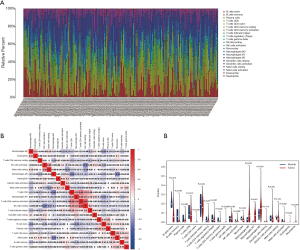
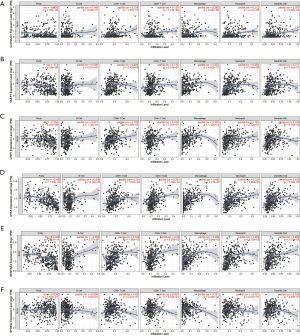
The results of the immune cell abundance profile of individual BC samples are shown in Figure 9A. The relationship between various immune cells and other immune cells showed that macrophages (M0), neutrophils, and resting mast cells had a negative relationship with naïve B cells and T-cell CD4 memory activation (Figure 9B). Compared to tumor samples, normal samples had significantly higher naïve B cells, B cell memory, and monocyte cell infiltration, but significantly lower macrophage (M0) infiltration (Figure 9C). The prognostic role of immune cells for BC showed that higher naïve B cells and T-cell CD4 memory activation had a better prognosis for BC, but higher infiltration of macrophages (M0), resting mast cells, and neutrophils led to poor BC prognosis (Figure 10). The results showed that immune cells may have a significant influence on BC prognosis via the regulation of other immune cell infiltration. Further, we explored the immune infiltration of our risk model’s messenger RNA (mRNA), and the results showed that all mRNA were positive correlation with Macrophages, while MYLK, SERPINE2, and SPTBN2 had a negative correlation with B cell infiltration (Figure 11).
Discussion
With the exception of protein-coding gene mutations and aberrant expression, the mutations and dysregulation of ncRNAs, in particular lncRNA, appear to have an essential role in cancer’s progress and prognosis. Many genome-wide associated studies of tumor samples have found that lncRNAs are significantly related to multiple cancer types. The aberrant expression and mutations of lncRNA can enhance tumorigenesis and metastasis, but several lncRNAs can act as tumor suppressors. Combined with miRNAs and target genes, lncRNA-miRNA-mRNA can serve as a comprehensive network that regulates gene expression. It can also act as diagnostic and prognostic biomarkers, and even as therapeutic targets for various cancer types.
To date, several lncRNAs, such as Urothelial Cancer Associated 1 (UCA1), HOX Transcript antisense RNA (HOTAIR), and Imprinted Maternally Expressed Transcript (H19), have been observed in BC (26). The overexpression of UCA1 can promote chemoresistance via regulation of the Wnt signaling pathway, and it may act as a potential urine diagnostics biomarker for BC (26). Shang et al. found that HOTAIR was overexpressed in BC samples compared with normal samples, and the overexpression of HOTAIR enhances tumor cell proliferation and induces resistance to doxorubicin, and appears to be an adverse biomarker for BC (27). In their research, Ariel et al. found that H19 was overexpressed in BC patients, resulting in a greater risk of BC recurrence (28). The potential functions of lncRNAs in BC requires further investigation.
In the present study, we comprehensive analyzed the regulatory network comprising lncRNAs, miRNAs, and mRNAs in a TCGA BC cohort. In total, 188 DElncRNAs, 200 DEmiRNA, and 2,661 DEmRNA were selected to construct the ceRNA network. Finally, 6 DEmiRNAs (has-mir-18a-5p, has-mir-347b-5p, has-mir-376c-3p, has-mir-590-3p, has-mir-214-3p, and has-let-7c-5p), 5 DElncRNAs (AC093010.3, MAGI2-AS3, HCG11, LINC00958, and ACO74117), and 97 DEmRNA were used for further analysis.
Potential mechanism of lncRNAs in regulating tumor’s progress are under research; many lncRNAs regulate gene expression by acting as miRNA sponges (29). We used the StarBase database to explore the ceRNA regulatory network of BC, and the results showed that AC093010.3, MAGI2-AS3, LINC00958, HCG11, and ACO74117 interacted with has-mir-18a-5p, has-mir-347b-5p, has-mir-214-3p, has-mir-376c-3p, has-mir-590-3p, and has-let-7c-5p, respectively.
We further used the ceRNA network to construct a model for predicting the prognosis of BC patients. The model (total, test, training) indicated that a high risk of BC led to poor outcomes for BC patients. Further analysis of the risk factors and other clinical features of BC prognosis showed that risk scores were an independent factor for predicting BC prognosis. The overexpression of CACNA2D1, GULP1, LTBP1, MYLK, SERPINE2, and SPTBN2 led to poor BC prognosis, but the overexpression of hsa-miR-590-3p appeared to be a favorable biomarker. Interestingly, Hayashi et al. showed that the downregulation of GULP1 could lead to cisplatin resistance (30). This indicates that the overexpression of CULP1 may act as a protector for BC. The overexpression of CACNA2D1 led to radioresistance non-small cell lung cancer stem-like cells (31). Cai et al. found that LTBP1 was overexpressed in esophageal squamous cell carcinoma (ESCC), and that the overexpression of LTBP1 was a promotor for ESCC progression via EMT and cancer-associated fibroblast transformation (32). Zhong et al. revealed that circular RNA MYLK acts as a ceRNA to accelerate the process of BC via regulating the Vascular Endothelial Growth Factor AVEGFA/Vascular Endothelial Growth Factor receptor 2 signaling pathway (33). Several studies have indicated SERPINE2 as a promotor for cancer progression (34,35). Du et al. showed that the overexpression of has-mir-590-3p could enhance chemoradiotherapy sensitivity for colorectal cancer (36). Based on these studies, we can conclude that DElncRNAs, DEmiRNAs, and DEmRNAs play a key role in tumorigenesis, tumor progression, and therapeutic response.
We not only explored the prognostic role of DElncRNAs, DEmiRNAs, and DEmRNA but also investigated their underlying potential BP in BC. The results of the functional enrichment analysis demonstrated that DElncRNAs, DEmiRNAs, and DEmRNAs had several biological functions and were involved in several significant pathways, such as the negative regulation of protein phosphorylation, the notch signaling pathway, the MAPK signaling pathway, the Rap1 signaling pathway, the TGF-β signaling pathway, and the TNF signaling pathway. Some of these functions or signaling pathways have been approved that played the critical role in several cancer types (37-41).
In addition to the dysregulation of lncRNAs, miRNAs, and mRNAs, tumorigenesis, tumor progression, and therapeutic response are also significantly influenced by the tumor microenvironment. We explored the relationship between DEmRNA, which significantly affects BC prognosis, and immune cell infiltration. The results showed that naïve B cells and T-cell CD4 memory activation were tumor suppressors, but macrophages (M0), resting mast cells, and neutrophils as tumor promotors for BC. There are significant differences in immune cell infiltrates between normal and tumor samples. Of these, DEmRNAs influenced BC prognosis via the regulation immune cell infiltration in BC. At steady state, the immune system can recognize and kill specific molecules expressed on tumor cells membranes, these up-expressed or down-expressed heterogeneous molecules can activate and increase the immune cells infiltration in the tumor microenvironment (42). In our study, up-regulation of SPTBN2 in bladder cancer can increased the abundance of macrophages cells and lead to poor prognosis of BC patients. A down-regulation of SERPINE2, CACNA2D1, GULP1, LTBP1, and MYLK in bladder cancer can reduced the abundance of macrophages cells and lead to poor prognosis of BC patients, and reduced the abundance of T-cell CD4 memory activation while lead to better prognosis. Expression of mRNAs in bladder cancer was altered via the ceRNA mechanism, and then influenced the immune cells infiltration in tumor microenvironment. The combined effect of these regulatory mechanisms leads to the dynamic development of bladder cancer tumors.
The limitation of our study is that absence of validation by experiments.
Conclusions
We constructed a lncRNA-miRNA-mRNA regulatory network by selecting DElncRNAs, DEmiRNAs, and mRNAs via comprehensive bioinformatics analysis. We found that CACNA2D1, GULP1, LTBP1, MYLK, SERPINE2, SPTBN2, and has-miR-590-3p could act as prognostic and diagnostic biomarkers for BC. LncRNA-HCG11 may influence BC via interacting with has-miR-590-3p and has-mir-376c-3p. Further studies are needed to determine the role of regulatory modules in the tumorigenesis, tumor progression, and therapeutic response of BC.
Acknowledgments
Funding: The present study was funded by the National Natural Science Foundation (No. 81600541).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-21-81
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-21-81). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Humphrey PA, Moch H, Cubilla AL, et al. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part B: Prostate and Bladder Tumours. Eur Urol 2016;70:106-19. [Crossref] [PubMed]
- Bresalier RS, Kopetz S, Brenner DE. Blood-based tests for colorectal cancer screening: do they threaten the survival of the FIT test? Dig Dis Sci 2015;60:664-71. [Crossref] [PubMed]
- Chamie K, Saigal CS, Lai J, et al. Quality of care in patients with bladder cancer: a case report? Cancer 2012;118:1412-21. [Crossref] [PubMed]
- Racioppi M, D'Agostino D, Totaro A, et al. Value of current chemotherapy and surgery in advanced and metastatic bladder cancer. Urol Int 2012;88:249-58. [Crossref] [PubMed]
- Glassman ML, de Groot N, Hochberg A. Relaxation of imprinting in carcinogenesis. Cancer Genet Cytogenet 1996;89:69-73. [Crossref] [PubMed]
- Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009;106:11667-72. [Crossref] [PubMed]
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223-7. [Crossref] [PubMed]
- Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta 2015;1856:151-64. [PubMed]
- Sanfilippo PG, Hewitt AW. Translating the ENCyclopedia Of DNA Elements Project findings to the clinic: ENCODE's implications for eye disease. Clin Exp Ophthalmol 2014;42:78-83. [Crossref] [PubMed]
- Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem 2014;9:1932-56. [Crossref] [PubMed]
- Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 2011;29:742-9. [Crossref] [PubMed]
- Walsh AL, Tuzova AV, Bolton EM, et al. Long noncoding RNAs and prostate carcinogenesis: the missing 'linc'? Trends Mol Med 2014;20:428-36. [Crossref] [PubMed]
- Askarian-Amiri ME, Crawford J, French JD, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 2011;17:878-91. [Crossref] [PubMed]
- Weber DG, Johnen G, Casjens S, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes 2013;6:518. [Crossref] [PubMed]
- Xue Y, Gu D, Ma G, et al. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis 2015;30:303-10. [Crossref] [PubMed]
- Ying L, Huang Y, Chen H, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst 2013;9:407-11. [Crossref] [PubMed]
- Zhang X, Gejman R, Mahta A, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res 2010;70:2350-8. [Crossref] [PubMed]
- Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 2010;42:1113-7. [Crossref] [PubMed]
- Ruff SM, Ayabe RI, Malekzadeh P, et al. MicroRNA-210 May Be a Preoperative Biomarker of Malignant Pheochromocytomas and Paragangliomas. J Surg Res 2019;243:1-7. [Crossref] [PubMed]
- Zhang H, Zhu M, Shan X, et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 2019;687:246-54. [Crossref] [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9. [Crossref] [PubMed]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature 2012;482:347-55. [Crossref] [PubMed]
- Si W, Shen J, Zheng H, et al. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics 2019;11:25. [Crossref] [PubMed]
- Lyu L, Xiang W, Zhu JY, et al. Integrative analysis of the lncRNA-associated ceRNA network reveals lncRNAs as potential prognostic biomarkers in human muscle-invasive bladder cancer. Cancer Manag Res 2019;11:6061-77. [Crossref] [PubMed]
- Wang X, Ding Y, Wang J, et al. Identification of the Key Factors Related to Bladder Cancer by lncRNA-miRNA-mRNA Three-Layer Network. Front Genet 2020;10:1398. [Crossref] [PubMed]
- Fan Y, Shen B, Tan M, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J 2014;281:1750-8. [Crossref] [PubMed]
- Shang C, Guo Y, Zhang H, et al. Long noncoding RNA HOTAIR is a prognostic biomarker and inhibits chemosensitivity to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother Pharmacol 2016;77:507-13. [Crossref] [PubMed]
- Ariel I, Sughayer M, Fellig Y, et al. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol 2000;53:320-3. [Crossref] [PubMed]
- Pennisi E. Shining a light on the genome's 'dark matter'. Science 2010;330:1614. [Crossref] [PubMed]
- Hayashi M, Guida E, Inokawa Y, et al. GULP1 regulates the NRF2-KEAP1 signaling axis in urothelial carcinoma. Sci Signal 2020;13:eaba0443 [Crossref] [PubMed]
- Sui X, Geng JH, Li YH, et al. Calcium channel alpha2delta1 subunit (CACNA2D1) enhances radioresistance in cancer stem-like cells in non-small cell lung cancer cell lines. Cancer Manag Res 2018;10:5009-18. [Crossref] [PubMed]
- Cai R, Wang P, Zhao X, et al. LTBP1 promotes esophageal squamous cell carcinoma progression through epithelial-mesenchymal transition and cancer-associated fibroblasts transformation. J Transl Med 2020;18:139. [Crossref] [PubMed]
- Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 2017;403:305-17. [Crossref] [PubMed]
- Liu J, Song S, Lin S, et al. Circ-SERPINE2 promotes the development of gastric carcinoma by sponging miR-375 and modulating YWHAZ. Cell Prolif 2019;52:e12648 [Crossref] [PubMed]
- Zhang J, Luo A, Huang F, et al. SERPINE2 promotes esophageal squamous cell carcinoma metastasis by activating BMP4. Cancer Lett 2020;469:390-8. [Crossref] [PubMed]
- Du B, Wang T, Yang X, et al. SOX9, miR-495, miR-590-3p, and miR-320d were identified as chemoradiotherapy-sensitive genes and miRNAs in colorectal cancer patients based on a microarray dataset. Neoplasma 2019;66:8-19. [Crossref] [PubMed]
- Anbalagan M, Rowan BG. Estrogen receptor alpha phosphorylation and its functional impact in human breast cancer. Mol Cell Endocrinol 2015;418:264-72. [Crossref] [PubMed]
- Balkwill F. TNF-alpha in promotion and progression of cancer. Cancer Metastasis Rev 2006;25:409-16. [Crossref] [PubMed]
- Colak S, Ten Dijke P. Targeting TGF-beta Signaling in Cancer. Trends Cancer 2017;3:56-71. [Crossref] [PubMed]
- Kontomanolis EN, Kalagasidou S, Pouliliou S, et al. The Notch Pathway in Breast Cancer Progression. ScientificWorldJournal 2018;2018:2415489 [Crossref] [PubMed]
- Lee S, Rauch J, Kolch W. Targeting MAPK Signaling in Cancer: Mechanisms of Drug Resistance and Sensitivity. Int J Mol Sci 2020;21:1102. [Crossref] [PubMed]
- Schneider AK, Chevalier MF, Derre L. The multifaceted immune regulation of bladder cancer. Nat Rev Urol 2019;16:613-30. [Crossref] [PubMed]