
Identification and validation of a prognostic immune-related lncRNA signature in bladder cancer
Introduction
Bladder cancer (Bca) was ranked fourth among malignant tumors in males in the United States in 2019 and eighth in the number of deaths (1). Cases of Bca can be classified as muscle-invasive bladder cancer (MIBC) and non-MIBC (NMIBC) based on the degree of tumor invasion. Approximately 20% of Bca cases are diagnosed as MIBC, and the rates of recurrence, progression, and mortality for MIBC are very high. Therefore, a variety of treatment modalities are used to treat MIBC (2,3). All stages of urothelial cancer patients’ 5-year OS rate is approximately 15–20% (4). Although the mechanisms regulating the occurrence and development of Bca have not been fully elucidated, recent in-depth studies of immune-related mechanisms, immunotherapies, and the application of immune-checkpoint inhibitors have led to the development of more effective treatment options for Bca (2-4). In the past few years, immunotherapy has become an important means of cancer therapy. Anti-CTLA-4 and Anti-PD-L1/PD1 antibodies have already demonstrated substantial clinical effects against a variety of solid tumors including Bca (2). Nevertheless, as the majority of patients with all stages of Bca do not respond to immunotherapy, there is a great need for novel immune-related therapeutic targets and additional biomarkers related to tumor prognosis.
Immune cells have been shown to interfere with molecular signaling and play a vital part in biological characteristics of cancers, and they are tightly associated with tumor invasion, proliferation, and metastasis (5). Studies have shown that an increase in tumor-infiltrating immune cells can be used as a predictive biomarker in various malignancies (6). It has also been reported that the balance between T cells and myeloid-derived suppressor cells, which is associated with skewing toward type 2 immunity, may influence the survival of patients with MIBC and predict Bca recurrence (7). Studies have shown that Bca plays an important role in promoting the accumulation of immunosuppressed myeloid cells and the inactivation of tumor-infiltrating lymphocytes. In addition, Bca was reported to be related to the high levels of regulatory T cells (Tregs). In addition, Infiltration of Tregs may promote bladder tumor invasion (8). Given the critical role of the immune system in cancer, to improve treatment and prognosis, it is necessary to identify all the immune-related factors involved in Bca.
Long non-coding RNAs (lncRNAs) are defined as non-protein-coding RNA transcripts that are longer than 200 nt (9). LncRNAs play key roles in regulating chromatin dynamics, gene expression, growth, differentiation, and development, and their dysregulation is associated with various types of cancer. Abnormal expression and mutation of lncRNAs can promote tumor development and metastasis (10). He et al. reported that the lncRNA LINC00958 is carcinogenic and promotes tumor metastasis in Bca (11). Studies have shown that numerous lncRNAs are widely expressed during T cell activation, differentiation and the activation of innate immune response. These lncRNAs play fundamental roles in regulating important aspects of immunity, such as inflammatory mediator production, cell differentiation, and cell migration (12). LncRNA LINK-A functions as an oncogene which plays a significant role in intrinsic tumor suppression and the presentation of cancer cell antigen (13). In hepatocellular carcinoma, lnc-EGFR acts as an immunosuppressor by promoting regulatory T cell differentiation (14). Nevertheless, the specific mechanism of immune-related lncRNAs in Bca is unclear. With the rapid development of immunotherapies, gene expression databases have been widely used to identify and explore potentially valuable therapeutic genes. In present research, immune-related lncRNAs of Bca were explored by using gene expression profiles with an aim to identify lncRNAs that have potential as prognostic indicators and immune therapeutic targets. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1353).
Methods
Publicly available datasets and immune-related genes
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Clinical data from patients with Bca were obtained from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). Samples lacking necessary information were excluded. Patients without survival data and those whose OS ≤30 days were excluded because they likely died due to non-neoplastic factors (i.e., severe infection and hemorrhage) rather than Bca. In total, 393 Bca patients were enrolled in this study. These patients were randomly divided into training and validation cohorts (ratio 7:3) for building and validating the prediction model, respectively. Computer-generated random number sequences were used for randomization. The clinicopathological characteristics of the Bca patients in the TCGA dataset were listed in Table S1. The immune-related genes (IRGs) were obtained from The Immunology Database and Analysis Portal (ImmPort, https://immport.niaid.nih.gov), and 1,300 IRGs were extracted. The clinical information for the Bca patients and the messenger RNA (mRNA) data are publicly available. All microarray data and RNA sequence data were standardized and log2 transformed with the R package.
Selection of lncRNA candidates and construction of an immune-related signature
Selection of lncRNAs was done according to Ensembl ID and annotation information. After construction of immune-related lncRNA coexpression networks (P<0.001), 1,249 lncRNAs were eligible for subsequent research. We then constructed a scale-free coexpression network using the weighted gene co-expression network analysis (WGCNA) package in R and the expression data for 1,249 immune-related lncRNAs in the training set.
Firstly, a hierarchical clustering analysis of Bca samples with different clinical characteristics was carried out based on immune-related lncRNAs expression to recognize outlier samples. Then, the lowest power at which the scale-free topology fit index reaches 0.8 was chosen to be the soft power (β) for construction of networks. In this study, β=3 was used to construct the scale-free network. Next, the adjacency was used to calculate the topological overlap matrix (TOM). We assigned immune-related lncRNAs into different modules, with a min-Module size of 30, by using hierarchical clustering based on the distance measure, the corresponding dissimilarity (1-TOM). Unassigned genes were classified as gray modules. Next, we defined two parameters: module eigengenes (MEs) and gene significance (GS), and based on them, we discovered modules that were markedly related to various clinical features of the patients with Bca. Among the non-gray modules, candidate modules which had the highest absolute correlation with OS were selected for further analysis. We used univariate Cox regression analysis to identify possible prognostic immune-related lncRNAs among the candidate modules. We further constructed a prognostic risk model and the formula for risk score was as follows:
where i is the number of prognostic genes, and βi and Expi represent regression coefficients and expression value, respectively.
After calculating the risk scores of the Bca patients, we divided them into low- and high-risk groups. Predictive power of the model was evaluated by the receiver operating characteristic (ROC) analysis. Kaplan-Meier curve analysis was performed to evaluate the significance of differences in OS between the high-risk and low-risk groups.
Construction of the nomogram
A nomogram model that integrates prognostic clinicopathological features can be used to assist clinical procedures and improve risk stratification. Here, we used clinical factors [age, sex, grade, stage, and tumor-node-metastasis (TNM) staging] and risk score of our model to build a prognostic nomogram to predict 1-, 3-, and 5-year OS in Bca patients with the “rms” package in R. A calibration plot and time-dependent ROC curve were generated to determine the calibration and identification of the model.
Functional enrichment analysis
DAVID 6.8 software was used to conduct KEGG pathway and GO annotation analyses based on genes from the candidate module. We performed principal component analysis (PCA) to discriminate sample groups and used gene set enrichment analysis (GSEA) to understand the underlying mechanism between different risk groups.
Statistical analysis
Univariate Cox regression analysis and Pearson correlation analysis were used to identify immune-related lncRNAs. Prognostic value of various parameters for OS was assessed by the Cox (proportional hazards) regression. For the establishment of a prognostic model, least absolute shrinkage and selection operator (LASSO) regression and multivariate Cox regression analyses were performed. Kaplan-Meier curves were created to compare the survival outcome. The ROC curve was plotted using the “survivalROC” package. The unpaired Student’s t-test, one-way analysis of variance (ANOVA), the Chi-square test, or Fisher’s exact test was used for analysis as appropriate. Statistical analyses were performed using R software 3.5.0 and IBM SPSS statistics version 22.0 (SPSS Inc., Chicago, IL, USA). A two-sided P value less than 0.05 was accepted as significant.
Results
Identification of immune-related lncRNAs
Totally, we obtained 1,300 IRGs from Immunology Database and Analysis Portal (ImmPort, https://immport.niaid.nih.gov). We extracted the sequence data and identified 14,152 lncRNAs from the dataset. Then, by constructing immune-lncRNA co-expression networks, we identified 1,249 immune-related lncRNAs (P<0.001).
Construct an immune-related signature to predict OS
We used the WGCNA tool with clinical traits (OS, grade, and stage) and RNA-seq data in the training set (Figure 1A). In total, we used one-step network construction method to obtain four modules, wherein β was equal to 3 (Figure 1B). Next, we performed a relevance analysis between the corresponding modules and clinical features. These outcomes revealed one module not only had the most significant correlation with OS but also had a significant correlation with grade and stage (yellow module; Figure 1C). Moreover, the scatter of OS or average GS in the module is shown in Figure 1D, and the yellow module was closely related to OS. Consequently, 239 genes were selected from the yellow module and the univariate Cox regression were used to further analysis. As shown in Figure 1E, 53 genes (53 protective genes and 0 risk genes) were significantly associated with OS (P<0.01). Then, in order to generate a prognostic signature in Bca cohort based on these 53 prognostic genes, multivariate Cox regression and LASSO Cox regression analyses were performed (Figure 1F,G). Ultimately, we constructed the signature based on the six genes (AC005674.2, AC090948.1, TFAP2A-AS1, AL354919.2, AC011468.1, and AC018809.2). As shown in Figure 1G, in the signature, the scatter of the regression coefficients was displayed.
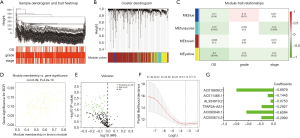
The immune-related gene signature is an independent prognostic factor for Bca
As shown in Figure 2, in the training cohort, the risk scores of the patients with Bca were ranked. And the risk scores of surviving patients were distinctly lower than those patients who died (P<0.0001). Bca patients in the low-risk group showed obviously better overall survival (OS) than those in the high-risk group (P<0.0001). The risk score could serve as an independent prognostic factor in Bca via multivariate cox regression analysis (HR =2.843, P<0.001). In addition, ROC analysis demonstrated that risk score was a prognostic marker of Bca, and the average AUC value was 0.707 (0.622–0.785, P<0.001) after 5 years of follow-up. In order to verify the dependability of the above conclusions, we further carried out the same analysis in the verification cohort.

The results showed that the risk scores of surviving patients were lower than those of patients who died in the validation set (P=0.0303). According to the Kaplan-Meier analysis, compared with high-risk patients, low-risk patients had significantly better OS (P<0.0001). In the validation set, the risk score remained an independent prognostic factor for Bca, and this result was consistent with the training set (HR =3.832, P<0.001). Moreover, the risk score had a powerful predictive value in the validation set, with an average AUC of 0.759 (0.624–0.884, P<0.001). Compared with other clinical indicators, the ROC value of the risk score in the validation and trainings set was the highest, indicating that the risk score is a reliable independent prognostic factor.
Clinicopathological characteristics of the low- and high-risk groups
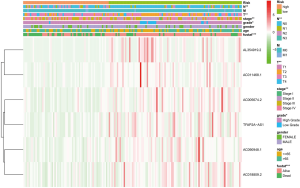
As shown in Figure 3, the heat map displayed six selected immune-related lncRNAs expression and clinicopathological characteristics in both high-risk and low-risk groups. We found dramatic differences between two groups in respect of grade (P<0.01), stage (P<0.05), and fustat (P<0.001; Figure 3).
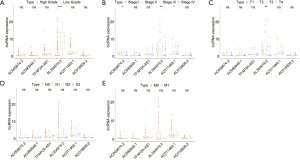
Next, we evaluated the expression of the six immune-related lncRNAs in Bca samples of different grades, stages, and TNM staging. The results revealed that the expression of lncRNA AL354919.2 was higher in lower grade tumors than in higher grade tumors (P<0.001), whereas other lncRNAs showed no significance differences with grade (Figure 4A). The expression of lncRNAs AC005674.2, AC011468.1, and AL354919.2 differed significantly with stage (P<0.05; P<0.05; P<0.01), while the other lncRNAs did not differ significantly with stage (Figure 4B). LncRNA AL354919.2 expression levels were associated with T stage (P<0.01) (Figure 4C). Notably, the expression levels of the six selected immune-related lncRNAs were not significantly associated with N and M stage (P<0.05; Figure 4D,E).
Construct nomogram for prognosis of Bca
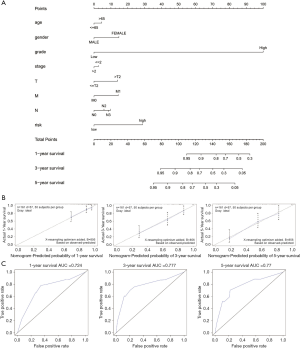
In order to set up a suitable method for monitoring prognosis of patients with Bca, we combined clinicopathological characteristics (age, sex, grade, stage, and TNM staging) and risk score to construct a prognostic nomogram. Analysis revealed that the prognostic nomogram could better predict the OS of Bca patients at 1, 3, and 5 years (Figure 5).
Functional enrichment analysis
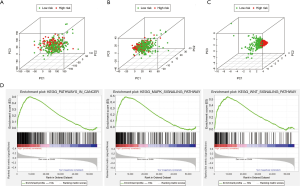
To explore differences between high-risk and low-risk groups according to the expression profiles of IRGs, risk genes, and all genes (Figure 6A,B,C), PCA method was performed in this study(Figure 6A,B,C). However, the immune status of two groups displayed no significant difference when the all genes’ expression profiles were included in the analysis (Figure 6A). The low- and high-risk groups were differentiated by immune status when PCA was performed using the immune gene expression profiles (Figure 6B). The risk genes were distributed in different directions, demonstrating that immune response of patients with Bca in high-risk group was quite different from those patients in low-risk group (Figure 6C). The results of a KEGG analysis indicated that these genes were associated with various immune-related signaling pathways, such as the T cell receptor, Toll-like receptor (TLR), B cell receptor, and JAK-STAT signaling pathways (Table S2). GO analysis revealed that the genes selected from the module were enriched in the biological process of tumor immunity, including regulation of the Wnt signaling pathway, MHC protein complex, cytokine receptor activity, and cytokine binding (Table S3). We further validated the functional annotation using GSEA, and the above outcomes indicated that between the two groups differentially expressed genes were enriched in some immune-related pathways and biological processes, such as pathways in cancer and the MAPK and WNT signaling pathways (Figure 6D).
Discussion
Bca is one of the most immunogenic tumors, and its somatic mutation rate ranks third among cancers. Bca can evade immune-mediated elimination, even if in the case of antigen-specific immune cell infiltration, which leads to challenges in the clinical treatment of Bca (8). Accumulating studies have demonstrated that tumor microenvironment plays a vital part in the progression and development of cancers, and a dysfunctional immune state in the tumor microenvironment is a hallmark of cancer. In tumor immunoregulation, immune system plays a dynamic role. And recent studies have reported that lncRNAs play fundamental regulatory roles in various immune system processes, including immune cell differentiation and function, antigen release and presentation, immune activation, and immune cell migration (13,15,16). Therefore, IR-lncRNAs can be used as a potential target for tumor immunotherapy, which is valuable for evaluating the survival and prognosis of tumor patients (6,13).
In this study, a six gene signature was constructed by using IR-lncRNAs in the training cohort and further verified it in validation cohort. To identify prospective immune-related lncRNAs that are involved in Bca, we used a dataset of 393 samples. WGCNA was performed to identify
We performed WGCNA to identify gene module closely associate with OS, and multivariate, univariate Cox regression, and LASSO analyses were utilized to establish an IR-lncRNAs gene signature from the candidate module. The Bca patients were divided into low-risk and high-risk groups. Patients in high-risk group displayed worse OS than patients in low-risk group both in the training and validation cohorts (P<0.0001). Our findings indicate that the risk score is closely related to malignant progression and poor prognosis. Six immune-related lncRNAs, AC005674.2, AC090948.1, TFAP2A-AS1, AL354919.2, AC011468.1, and AC018809.2, were considered to be prognostic factors for Bca based on their risk scores. ROC analysis showed that both in the training and validation cohorts, risk score had strong prognostic ability. We noted that risk score is an important predictor both in two sets, the mean AUC of 0.707–0.759. The risk scores with clinical feature was combined and a nomogram based on the two cohorts was constructed to ameliorate risk stratification and satisfy clinical need. The 1-, 3-, and 5-year OS of Bca patients could be predicted by the nomogram. ROC analysis indicated that the accuracy of prognosis was obviously enhanced by combining clinical parameters with risk score, and the average AUC value was 0.77 after 5 years of follow-up. Furthermore, the calibration plot showed good consistency between the predicted results and the observed results.
Immunotherapy has rapidly changed the treatment course of various cancers, including renal carcinoma and lung cancer; it also has become a new strategy for the treatment of patients with Bca, particularly those with cancer that could not be controlled by first-line chemotherapy (17,18). All six immune-related lncRNAs (AC005674.2, AC090948.1, TFAP2A-AS1, AL354919.2, AC011468.1, and AC018809.2) in our signature were protective genes. Some of them were reported to be enrolled in multiple tumors. The present study suggested that TFAP2A-AS1 may serve as a tumor suppressor, and in breast cancer it was previously shown to be related to better prognosis. As was reported that TFAP2A-AS1 functioned as a miR-933 sponge, which degrades smad2 mRNA, and showed inhibitory effect on breast cancer cell invasion and proliferation (19). However, Jiang et al. reported that TFAP2A-AS1 high expression was associated with poor prognosis in clear cell renal cell carcinoma patients (20). We found that TFAP2A-AS1 was a protective factor for Bca, and the specific mechanism underlying the function of this gene in Bca requires further study. The mechanisms of the other five lncRNAs also have not been reported and thus require further study. It was worth noting that of the six analyzed immune-related lncRNAs, only AL354919.2 was found to be related to Bca grade and stage, suggesting that this gene is closely related to Bca prognosis and warrants further research.
In the bladder tumor microenvironment, various interconnected intrinsic and extrinsic immunoregulatory pathways have been identified, revealing new predictive tools and therapeutic targets (21). In our study, a signature comprising six immune-related lncRNAs was enrolled in the regulation of immune response of Bca.
According to functional enrichment analysis, the genes in candidate modules were enriched in several immune-related pathways, such as the JAK-STAT, TLR, and WNT signaling pathways. The JAK-STAT signaling pathway has been reported to mediate various immunomodulatory processes, including tumor-driven immune escape and tumor cell recognition (22). TLRs play crucial roles in both innate immunity and cancer therapy (23). The GSEA results showed that in high-risk group several immune-related pathways were enriched. We also found that the WNT signaling pathway was enriched in both analyses. Notably, the WNT signaling pathway can impact not only the biology of (pre)malignant cells but also the ability of cancer cells to evade the immune system (24). Thus, we inferred that signaling pathway of WNT might play significant parts in Bca, simultaneously may be closely related to tumor immunity.
The results of our research provide possible targets for immunotherapy and markers for the prognostic evaluation of Bca and thus may improve the clinical efficacy of Bca treatment. Herein, we conducted a preliminary exploration of immune-related lncRNAs in Bca. However, our study had some limitations. In follow-up studies, we need to increase the sample size and verify the observed characteristics of additional Bca cases. In addition, the functions and mechanisms of these genes require further study. For clinical application, these findings need to be further validated by immunohistochemistry or flow cytometry. Despite these limitations, we described a signature of six immune-related lncRNAs that has not been studied previously and is tightly associated with the risk score, tumor status, immune response, and OS of Bca patients.
To conclude, we identified six immune-related lncRNAs that have prognostic value for Bca patients. Based on the median risk score, patients with Bca were separated into low-risk and high-risk groups, and they had different immune statuses. We hope that these immune-related lncRNAs will be useful as treatment targets and prognostic markers of Bca and provide new strategies for Bca immunotherapy.
Acknowledgments
We are grateful for having had free access to the TCGA databases. Lori from Editage helped to edit the language of this paper.
Funding: This work was funded and supported by the National Natural Science Foundation of China (No. 81871151) and the Class A of Major Medical Talents in the Jiangsu Province (No. ZDRCA2016009).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1353
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1353). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Crispen PL, Kusmartsev S. Mechanisms of immune evasion in bladder cancer. Cancer Immunol Immunother 2020;69:3-14. [Crossref] [PubMed]
- Pettenati C, Ingersoll MA. Mechanisms of BCG immunotherapy and its outlook for bladder cancer. Nat Rev Urol 2018;15:615-25. [Crossref] [PubMed]
- Aggen DH, Drake CG. Biomarkers for immunotherapy in bladder cancer: a moving target. J Immunother Cancer 2017;5:94. [Crossref] [PubMed]
- Wei C, Liang Q, Li X, et al. Bioinformatics profiling utilized a nine immune-related long noncoding RNA signature as a prognostic target for pancreatic cancer. J Cell Biochem 2019;120:14916-27. [Crossref] [PubMed]
- Darvin P, Toor SM, Sasidharan Nair V, et al. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 2018;50:1-11. [Crossref] [PubMed]
- Chevalier M F, Trabanelli S, Racle J, et al. ILC2-modulated T cell-to-MDSC balance is associated with bladder cancer recurrence. J Clin Invest 2017;127:2916-29. [Crossref] [PubMed]
- Smith SG, Zaharoff DA. Future directions in bladder cancer immunotherapy: towards adaptive immunity. Immunotherapy 2016;8:351-65. [Crossref] [PubMed]
- Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol 2017;35:177-98. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- He W, Zhong G, Jiang N, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest 2018;128:861-75. [Crossref] [PubMed]
- Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol 2014;35:408-19. [Crossref] [PubMed]
- Li Y, Jiang T, Zhou W, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun 2020;11:1000. [Crossref] [PubMed]
- Jiang R, Tang J, Chen Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun 2017;8:15129. [Crossref] [PubMed]
- Yu WD, Wang H, He QF, et al. Long noncoding RNAs in cancer-immunity cycle. J Cell Physiol 2018;233:6518-23. [Crossref] [PubMed]
- Denaro N, Merlano MC, Lo Nigro C. Long noncoding RNAs as regulators of cancer immunity. Mol Oncol 2019;13:61-73. [Crossref] [PubMed]
- Butt SU, Malik L. Role of immunotherapy in bladder cancer: past, present and future. Cancer Chemother Pharmacol 2018;81:629-45. [Crossref] [PubMed]
- Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet 2016;388:2796-810. [Crossref] [PubMed]
- Zhou B, Guo H, Tang J. Long Non-Coding RNA TFAP2A-AS1 Inhibits Cell Proliferation and Invasion in Breast Cancer via miR-933/SMAD2. Med Sci Monit 2019;25:1242-53. [Crossref] [PubMed]
- Jiang W, Guo Q, Wang C, et al. A nomogram based on 9-lncRNAs signature for improving prognostic prediction of clear cell renal cell carcinoma. Cancer Cell Int 2019;19:208. [Crossref] [PubMed]
- Schneider AK, Chevalier MF, Derré L. The multifaceted immune regulation of bladder cancer. Nat Rev Urol 2019;16:613-30. [Crossref] [PubMed]
- Owen KL, Brockwell NK, Parker BS. JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression. Cancers (Basel) 2019;11:2002. [Crossref] [PubMed]
- Braunstein MJ, Kucharczyk J, Adams S. Targeting Toll-Like Receptors for Cancer Therapy. Target Oncol 2018;13:583-98. [Crossref] [PubMed]
- Galluzzi L, Spranger S, Fuchs E, et al. WNT Signaling in Cancer Immunosurveillance. Trends Cell Biol 2019;29:44-65. [Crossref] [PubMed]


