
Downregulation of PARVA promotes metastasis by modulating integrin-linked kinase activity and regulating MAPK/ERK and MLC2 signaling in prostate cancer
Introduction
Prostate cancer (PCa) is the most commonly diagnosed malignancy for males worldwide and one of the main causes of cancer death in Western countries (1). Although localized PCa is highly curable, metastatic PCa remains invariably fatal, with the 5-year survival rates being lower than 30% (2). The development and progression to metastasis is a pivotal event correlating with patient outcomes and the availability of therapeutic options. Therefore, unravelling the molecular mechanisms underlying the progression to metastasis in PCa may substantially help identify novel targets for improving therapeutic effect and facilitating preventive interventions.
Extracellular matrix (ECM), integrin-related adhesion proteins, and actin cytoskeleton are critically involved in tumor cell invasion and metastasis (3). PARVA (α-parvin), also known as actopaxin or CH-ILKBP, is a focal adhesion protein that physically interacts with integrin-linked kinase (ILK) and cysteine-histidine-rich protein (PINCH) in the ILK-PINCH-Parvin (IPP) complex (4-6). The IPP complex associates with integrin heterodimers and therefore activates multiple extracellular signaling including Akt, extracellular signal-regulated kinase (ERK)1/2 and Rac1 small GTPase (7). Several studies have shown that PARVA contributes to alterations of cell morphology, ECM, migration, invasion, and survival processes (8-14). In addition, recent evidence identifies PARVA and miR-130b may form a pair that plays a key biological role that associate with PCa proliferation and metastasis. PARVA mRNA is downregulated in PCa samples and correlated with biochemical recurrence (BCR)-free survival (15). However, whether PARVA plays a role in PCa progression and the underlying molecular mechanism are still unknown.
In the present study, we determined the PARVA expression status in published microarray data and clinical PCa specimens. In vitro functional assays were applied to characterize the role of PARVA, and its molecular mechanism in PCa metastasis was further explored.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tau-21-108).
Methods
Clinical specimens
Paraffin-embedded specimens from primary (n=198) and lymph node metastatic (n=36) PCa tissues were harvested through the archives of the Institute of Urology, Peking University (IUPU, Beijing, China). The hematoxylin and eosin (HE) slides were reviewed and validated by two pathologists. Detained clinical and pathological characteristics of patients were obtained from the medical records. All procedures undertaken in our study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Board of Peking University First Hospital (2015-977). The requirement for written informed consents were waived because of the retrospective nature of the study.
Cell lines and cell culture
Human embryonic kidney (HEK293T), human normal prostate epithelial cell line (RWPE-1), human benign prostatic hyperplasia epithelial (BPH-1), and human PCa (LNCaP, C4-2, PC-3, 22RV1, and VCaP) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Mycoplasma contamination was routinely checked when cell growth or shape was changed. All the cell lines were cultured in RPMI-1640 or Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Corning, Corning, NY, USA) in a 5% CO2 humidified atmosphere at 37 °C.
Plasmids, retroviral infection, and transfection
The PARVA and ILK short hairpin RNA (shRNA) oligonucleotides were designed using an online shRNA designer tool (https://portals.broadinstitute.org/gpp/public/), followed by insertion into pLKO.1-TRC vector (Addgene, Watertown, MA, USA). The PARVA cDNA was amplified using PCR and then inserted into a pLVX-IRES-Bla vector (Addgene). The insertion was confirmed by DNA sequencing. The polyethylenimine (PEI) reagent (Polysciences Inc., Warrington, PA, USA) was used to transfect the plasmids into cells according to the manufacturer’s instructions. Stable knockdown or overexpression cell lines were selected for 2 weeks by treatment with 1.0 μg/mL of puromycin or 5 μg/mL of Blasticidin for 36 hours after infection. Transfection efficiency was confirmed by real-time polymerase chain reaction (RT-PCR) and western blot assay. The detailed sequences data are described in Table S1.
Bioinformatics analysis
Publicly available PCa microarray data sets of GSE21032 (16), GSE35988 (17), GSE55945 (18), GSE68882 (19), GSE8702 (20), GSE6919 (21,22), GSE3325 (23), GSE3933 (24,25), and GSE6099 (26) were downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). The Cancer Genome Atlas (TCGA, n=551) data set was downloaded from the web portal (https://cancergenome.nih.gov/). The expressed genes were subsequently analyzed for biological themes using gene set enrichment analysis (GSEA), and a formatted GCT file was used as input for the GSEA algorithm v2.0 (http://software.broadinstitute.org/gsea/index.jsp).
Quantitative RT-PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), and complement DNA (cDNA) was synthesized with the Reverse Transcription System (Roche, Basel, Switzerland) following the manufactures’ protocols. Quantitative RT-PCR was performed in triplicate on 96-well optical plates using Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific,) and SYBR Green PCR Master Mix (Roche) according to the manufacturer’s instructions. GAPDH and α-tubulin were used as the endogenous control. The primers used are described in Table S2.
Western blot analysis
Cell lysates were separated onto 10% sodium dodecyl sulfate (SDS)-polyacrylamide (8–12%) gels, and transferred on to polyvinylidene fluoride membrane (Millipore, Darmstadt, Germany). The membranes were blocked with Tris-buffered saline solution containing 5% skim milk and 0.1% Tween-20 for 1 hour at room temperature and incubated with a primary antibody overnight at 4 °C. The membranes were then incubated with appropriate horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 hours at room temperature, and finally visualized using the ECL Plus kit (Applygen Technologies Inc., Beijing, China). Immunoblots were quantified using the ImageJ program (National Institutes of Health (NIH), Bethesda, MD, USA). All antibodies used in this study are listed in Table S3.
Coimmunoprecipitation
C4-2 cells stably expressing S-tagged PARVA(S-PARVA) were lysed in NETN (150 mM NaCl, 1 mM EDTA, 50 mM Tris-Cl, pH 7.4–7.5, 1.0% NP-40) buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF), protease inhibitors, and phosphatase inhibitors (Roche). Samples were incubated for 30 minutes on ice and centrifuged at 12,000× g for 30 minutes at 4 °C. Supernatants were then incubated with S-protein beads (Millipore) overnight at 4 °C. The beads were washed 3 times with NETN buffer. Proteins were eluted from agarose beads with SDS sample buffer and subjected to SDS-PAGE. Proteins bands were excised and subjected to mass spectrometry analysis.
Immunohistochemistry
Immunohistochemical (IHC) staining was carried on the paraffin-embedded PCa tissues following the manufacturer’s protocol. The tissue slides were dried for 1 hour in an oven at 70 °C, dewaxed, rehydrated, and autoclaved in citrate retrieval buffer. Tissue sections were blocked with 0.3% hydrogen peroxide solution, and then incubated with rabbit polyclonal anti-PARVA antibody (1:500, cat no. 11202-1-AP, Proteintech Group, Rosemont, IL, USA) overnight at 4 °C, and with a biotinylated secondary antibody for 1 hour at room temperature. The immune complexes were visualized using the 3, 3’-diaminobenzidine tetrahydrochloride, and tissue slides were counterstained with hematoxylin. The tissue slides were reviewed and assessed blindly by two independent pathologists. The PARVA expression were quantified into 4 grades by staining intensity (0, no staining; 1, weak staining, light yellow; 2, moderate staining, yellow brown; 3, strong staining, brown) and the percentage of extent of reactivity (0, <5% positive cells; 1, 5–25% positive cells; 2, 26–50% positive cells; 3, 51–75% positive cells; 4, >75% positive cells). An immunohistochemical expression score was obtained by multiplying the intensity and reactivity extension values (range, 0–12).
In vitro proliferation, migration, and invasion assays
Cell proliferation was measured by CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS; Promega, WI, USA) according to the manufacturer’s instructions. Cells were seeded at 1,000 cells per well in 96-well plates with 6 replicates, and incubated with 100 μL/well of the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium reagent (Promega) for 3 hours. The absorbance at 490 nm was measured using a spectrophotometer (Thermo Fisher Scientific), and proliferation curves were constructed.
Cell migration and invasion assays were performed with Transwell inserts containing a polycarbonate membrane with 8.0 μm pores (Corning), and conducted with uncoated and coated Matrigel, respectively. Briefly, the indicated cells were seeded in the upper chambers containing 200 μL serum-free medium while the lower chambers contained 700 μL medium supplemented with 10% FBS. After 36 hours of incubation at 37 °C, the cells were fixed with 4% paraformaldehyde and stained with Crystal Violet (MilliporeSigma, St. Louis MO, USA). Cells on the upper surface of the membrane were gently removed with a cotton swab, and migrated or invaded cells on the lower surface of the membrane were counted under the microscope (Olympus, Tokyo, Japan).
Statistical analysis
All data were analyzed using GraphPad prism 8.0 (GraphPad, San Diego, CA USA) or SPSS 20.0 software (IBM, Armonk, NY, USA), and results are expressed as mean and standard deviation (SD). A two-tailed Student’s t-test was performed to compare the continuous variables. Correlation analysis was assessed using chi-square and Pearson’s correlation coefficient test. Survival rate analysis was performed by drawing and calculating the log-rank P test using the Kaplan-Meier method. Significance values were set at *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
Results
Expression of PARVA in primary and metastatic PCa
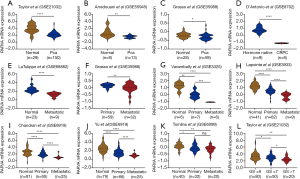
To identify whether any difference of PARVA expression exists between primary PCa, metastatic PCa, castration-resistant prostate cancer (CRPC), and noncancerous prostate tissues, we first analyzed the mRNA expression of PARVA in multiple public microarray data sets. Gene expression profile data from GSE21032, GSE55945, and GSE35988 showed that PARVA expression was lower in primary PCa tissues than that in normal adjacent benign prostate tissues (Figure 1A,B,C). The data from GSE8702 suggested that PARVA was downregulated in CRPC samples compared with hormone-naïve PCa samples (Figure 1D). Microarray data of GSE68882 and GSE35988 demonstrated that PARVA mRNA level was reduced in metastatic PCa specimens compared with primary PCa specimens (Figure 1E,F). Furthermore, the data from GSE3325, GSE3933, GSE6919, and GSE6909 indicated a gradual decrease in PARVA expression from noncancerous prostatic tissues to primary PCa tissues and metastatic PCa tissues (Figure 1G,H,I,J,K).
Downregulation of PARVA was associated with poor prognosis in PCa
To further investigate the clinicopathological relevance of PARVA expression in human PCa, we performed data analyses of GSE21032 and identified that PARVA was downregulated in patients with higher Gleason score relative to patients with lower Gleason score (Figure 1L). In TCGA cohort, we also found that PARVA mRNA expression was downregulated in primary PCa tissues compared to normal adjacent specimens (Figure 2A). PARVA expression was associated with Gleason score (Figure 2B) and lymph node status (Figure 2C). Notably, Kaplan-Meier survival analysis revealed that PCa patients with PARVA downregulation had a faster progression to BCR (Figure 2D).
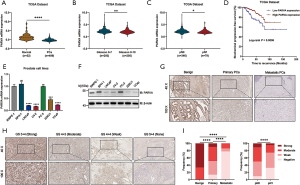
We then examined PARVA expression in a series of prostate cell lines (LNCaP, C4-2B, PC-3, 22RV1, benign prostatic hyperplasia cell line, BPH1, and normal prostate epithelial cell line, RWPE-1). The mRNA and protein expression of PARVA was lower in PCa cell lines than in RWPE-1 cell lines, except for the PC-3 cells (Figure 2E,F).
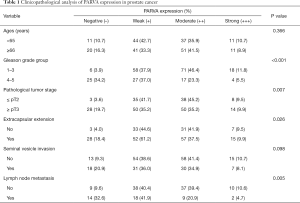
The clinical relevance of PARVA was further assessed by IHC staining in 198 primary PCa and 36 lymph node metastatic PCa samples. As shown in Figure 2G, the PARVA protein was downregulated in primary PCa tissues compared with adjacent benign tissues, with the lowest expression being observed in metastatic tumors. We also examined PARVA protein expression in primary PCa tissues and evaluated its correlation with clinicopathological characteristics. As shown in Table 1, PARVA downregulation in PCa was correlated with relatively higher Gleason score (P<0.001). The representative IHC images of PARVA in PCa tissues with different Gleason score are shown in Figure 2H. In addition, decreased PARVA levels were associated with pathological tumor stage (P=0.007), extracapsular extension (P=0.026), and lymph node metastasis (P=0.005; Figure 2I). There was no significant association between PARVA expression and the age of patients (P=0.366) or seminal vesicle invasion (P=0.098). Taken together, these results indicated that PARVA downregulation was associated with PCa progression and poor prognosis.
Full table
Knockdown of PARVA promoted migration and invasion of PCa cells
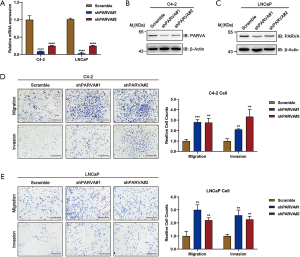
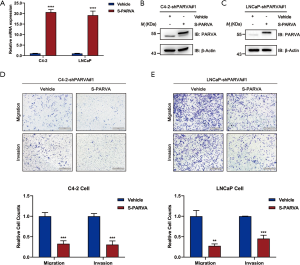
We used stable knockdown strategies mediated by shRNA in C4-2B and LNCaP cell lines. The efficiency of PARVA knockdown was confirmed by quantitative RT-PCR (Figure 3A) and western blotting (Figure 3B,C). Transwell assays demonstrated that knockdown of PARVA markedly increased migration and invasion in C4-2B (Figure 3D) and LNCaP (Figure 3E) cells.
However, we MTS assay revealed that PARVA downregulation mediated by shRNA would not affect cell proliferation (Figure S1A,B), and colony formation assay (Figure S1C). These experiments confirmed the metastasis-suppressive effect of PARVA in PCa.
Restoration of PARVA expression attenuated migration and invasion of PCa cell lines
In order to confirm the inhibitory effects of PARVA on PCa cell metastasis, expression of PARVA was restored in C4-2B and LNCaP cells. Overexpression effects were confirmed by quantitative RT-PCR (Figure 4A) and western blotting (Figure 4B,C). As shown in Figure 4D and E, restoration of PARVA expression attenuated the migratory and invasive abilities of C4-2B and LNCaP cells compared with control vehicle cells.
PARVA inhibited MAPK/ERK signaling in PCa cells
To investigate the molecular mechanisms via which PARVA loss promotes PCa metastasis, we performed S fusion protein-pull down experiments coupled with tandem mass spectrum analysis of PARVA-containing complexes in C4-2 cells and identified the number of proteins, with the top-ranked proteins being the well-known PARVA-associated proteins such as ILK (Figure 5A,B). Previous studies have shown that PARVA forms a ternary complex with ILK and PINCH, which is involved in a diverse set of signaling pathways (7). Increasing the ILK activity results in the phosphorylating Akt/PKB and GSK3β and then promotes epithelial-mesenchymal transition (EMT) which (27,28) is a crucial step for tumor relapse and metastasis (29). First, we confirmed with the coimmunoprecipitation assay that PARVA interacted with ILK in PCa cells (Figure 5C). We then investigated whether PARVA can affect Akt/PKB and GSK3β signaling. However, western blotting did not show that PARVA overexpression influenced EMT, Akt/PKB, or GSK3β pathways (Figure S2A,B).
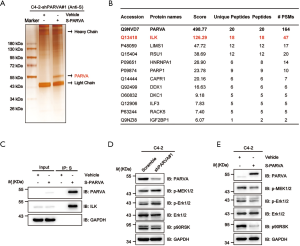
The MAPK/ERK signaling cascade is another pathway that is often aberrantly activated in advanced metastatic PCa (30). Several studies have reported that activation of ILK can also lead to upregulation of MAPK/ERK pathways (31-33). In the present study, we found that downregulation of PARVA increased the phosphorylation of ERK1/2 (Figure 5D) and conversely, PARVA overexpression inhibited activation of the MAPK/ERK signaling pathway (Figure 5E).
ILK was critically involved in the metastasis-suppressing function of PARVA
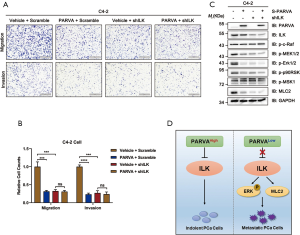
To further investigate whether PARVA-mediated phenotypes arise predominantly via ILK activation, we examined how blocking ILK signaling pathway affected the metastasis capability in C4-2 cells. The ILK was knocked down in the PARVA-overexpressing C4-2 cells using an interfering shRNA protocol. As shown in Figure 6A and B, the knockdown of ILK expression abrogated the suppressive migration and invasion induced by PARVA overexpression compared with the expression of scrambled control shRNA in C4-2 cells. With knockdown of ILK, the downregulated MAPK/ERK phosphorylation and Myosin Light Chain 2 (MLC2) expression by PARVA overexpression was abolished (Figure 6C). These results indicated that ILK is essential for the PARVA-mediated metastasis suppression in PCa cells (Figure 6D).
Discussion
The majority of patients with localized PCa can be treated by radical therapy which yields a 5-year survival rate of almost 100% (1). However, the disease usually becomes incurable once the tumor spreads out beyond the prostate and progresses to distant metastasis (34). Therefore, identification of the molecular alterations that drive PCa metastasis is crucial for understanding the underlying the metastatic mechanism and improving survival in patients with PCa. In this study, we attempted to characterize the role of PARVA in the development and progression of PCa. Previous studies have documented that PARVA is overexpressed in multiple solid malignancies including carcinoma of lung, breast, and other sites (13,35). However, Zhang et al. also reported that PARVA could negatively regulate lamellipodium formation in HeLa cells (36). Thus, the function of PARVA in diverse tumors is dual, and may depend on the organ-specific action and cellular microenvironment.
In the present study, we demonstrated that PARVA was significantly underexpressed in PCA among several cohorts. After analyzing the microarray data sets, we found that PARVA was downregulated in primary PCa relative to tissues adjacent to cancer, metastatic PCa relative to primary PCa, and castration-resistant PCA relative to hormone-naïve PCa. This indicated that PARVA might act as a suppressor gene in metastasis and castration-resistant-associated progression in PCa. Our results further suggested that PARVA downregulation was significantly correlated with adverse pathological characteristics and was an independent predictor for the poor BCR-free survival. Consistent with our findings, Aakula et al. recently have shown that PARVA, as a putative target of miR-130b, is downregulated in clinical PCa tissues through systematic functional screening, with this underexpression being significantly correlated with the BCR-free survival (15). In addition, the IHC analysis in our study demonstrated that PARVA protein underexpression was independently associated with unfavorable clinical outcome in patients with PCa. These results confirm the finding of our previous study, and to our best knowledge, this is the first direct evidence for the association of PARVA and the clinicopathological characteristics of PCa. However, larger-scale cohort studies may be required to identify whether PARVA is an effective biomarker of PCa prognosis.
It is well documented that ILK is the most potent mediator of PARVA in the regulation of tumor cell micromorphology, migration, invasion, and survival signaling (6,8-14). Dysregulation of ILK signaling was found in several hormonal tumors including breast, ovary and prostate (37). Hu et al. reported that ILK overexpression was associated with poorly-differentiated PCa (38). Aberrant ERG translocations activates ILK-mediated EMT progression in approximately half of PCa (39). Moreover, ILK inhibitors induces apoptosis and delays growth in PTEN-deficient PCa cells (40). Although a recently detailed sequence analysis suggested that ILK is a pseudokinase as it lacks several key active site residues, numerous studies have shown that ILK can act as a bona fide protein kinase, with the capacity to phosphorylate AKT, GSK3β, and other substrates (6,7,41). In several malignancies, PARVA overexpression has been shown to promote ILK activity, and then activate and inhibit its downstream mediators, AKT and GSK3β (7). In this study, the ectopic expression of PARVA did not affect AKT and GSK3β phosphorylation. Recently, Velazquez-Torres et al. also found that silencing of PARVA in melanoma cells did not influence AKT phosphorylation, which consistent with our results (42).
The MAPK/ERK signaling cascade is frequently activated aberrantly in advanced cancers, including metastatic PCa. Bluemn et al. found that FGF/MAPK pathway was notably elevated in metastatic PCa with an AR-null phenotype (43). Similarly, Gao et al. described the activation of AKT and ERK signaling in androgen-independent PCa (44). The MAPK/ERK pathway has also been reported to be one of the mediators of ILK in an array of malignant tumors (31,45). In our study, knockdown of PARVA increased the phosphorylation of ERK1/2, while the overexpression of PARVA reversed this phenomenon through inhibiting the MAPK/ERK signaling.
Previous studies have demonstrated that the interaction between ILK and PARVA, and the ILK-mediated phosphorylation of downstream substrates, were abolished via mutating ILK (10,46,47). In addition, Montanez et al. reported that the absence of PARVA/ILK impaired spreading, polarity, and migration via increasing RhoA and Rho-kinase (ROCK)-mediated signaling in vascular smooth muscle cells (12). A similar role for PARVA/ILK complex in regulating Rho activity has been observed in Schwann cells of the nervous system (48). These studies corporately identify the PARVA/ILK complex as a negative regulator of cell mobility in certain cell types, and PARVA as the key adaptor of this function.
Our study showed that knockdown of ILK expression significantly reversed the capacity for PARVA overexpression to suppress the migration and invasion of the C4-2 cells. In addition, the knockdown of ILK protein blocked the inhibitory effect of PARVA on phosphorylation of ERK1/2 and its downstream signaling. Collectively, our results suggested that underregulated PARVA in PCa might promote cancer progression through, at least in part, releasing the inhibition of ILK activity and, consequently upregulating the MAPK/ERK pathway. However, our study did not explore the more detailed relationship between PARVA and ILK, and the different mechanisms among different PARVA-ILK complexes in PCa still remain unclear. Further investigations were warranted in discussing the potent between PARVA and ILK are needed to explore these issues.
Acknowledgments
Funding: This study was supported by funding from the Peking University Medicine Seed Fund for Interdisciplinary Research (No. BMU2020MI003), the Capital Clinical Research Project of Beijing Municipal Science and Technology Commission (No. Z141107002514089), and the Wu Jieping Medical Fund (No. 320.6750.12273).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tau-21-108
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-21-108.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-21-108). Prof. GS serves as an unpaid editorial board member of Translational Andrology and Urology from Jun 2019 to May 2021. Prof. LZ serves as an unpaid editorial board member of Translational Andrology and Urology from Mar 2020 to Feb 2022. The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures undertaken in our study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Board of Peking University First Hospital. The requirement for written informed consents were waived because of the retrospective nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Cornford P, van den Bergh RCN, Briers E, et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer. Part II-2020 Update: Treatment of Relapsing and Metastatic Prostate Cancer. Eur Urol 2021;79:263-82. [Crossref] [PubMed]
- Kechagia JZ, Ivaska J, Roca-Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol 2019;20:457-73. [Crossref] [PubMed]
- Tu Y, Huang Y, Zhang Y, et al. A new focal adhesion protein that interacts with integrin-linked kinase and regulates cell adhesion and spreading. J Cell Biol 2001;153:585-98. [Crossref] [PubMed]
- Legate KR, Montanez E, Kudlacek O, et al. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat Rev Mol Cell Biol 2006;7:20-31. [Crossref] [PubMed]
- Wickström SA, Lange A, Montanez E, et al. The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! EMBO J 2010;29:281-91. [Crossref] [PubMed]
- Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, et al. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer 2010;10:858-70. [Crossref] [PubMed]
- Nikolopoulos SN, Turner CE. Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion. J Cell Biol 2000;151:1435-48. [Crossref] [PubMed]
- Nikolopoulos SN, Turner CE. Molecular dissection of actopaxin-integrin-linked kinase-Paxillin interactions and their role in subcellular localization. J Biol Chem 2002;277:1568-75. [Crossref] [PubMed]
- Huang AH, Pan SH, Chang WH, et al. PARVA promotes metastasis by modulating ILK signalling pathway in lung adenocarcinoma. PLoS One 2015;10:e0118530. [Crossref] [PubMed]
- LaLonde DP, Brown MC, Bouverat BP, et al. Actopaxin interacts with TESK1 to regulate cell spreading on fibronectin. J Biol Chem 2005;280:21680-8. [Crossref] [PubMed]
- Montanez E, Wickstrom SA, Altstatter J, et al. Alpha-parvin controls vascular mural cell recruitment to vessel wall by regulating RhoA/ROCK signalling. EMBO J 2009;28:3132-44. [Crossref] [PubMed]
- Ng L, Tung-Ping Poon R, Yau S, et al. Suppression of actopaxin impairs hepatocellular carcinoma metastasis through modulation of cell migration and invasion. Hepatology 2013;58:667-79. [Crossref] [PubMed]
- Fukuda K, Gupta S, Chen K, et al. The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Mol Cell 2009;36:819-30. [Crossref] [PubMed]
- Aakula A, Kohonen P, Leivonen SK, et al. Systematic Identification of MicroRNAs That Impact on Proliferation of Prostate Cancer Cells and Display Changed Expression in Tumor Tissue. Eur Urol 2016;69:1120-8. [Crossref] [PubMed]
- Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11-22. [Crossref] [PubMed]
- Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239-43. [Crossref] [PubMed]
- Arredouani MS, Lu B, Bhasin M, et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res 2009;15:5794-802. [Crossref] [PubMed]
- LaTulippe E, Satagopan J, Smith A, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res 2002;62:4499-506. [PubMed]
- D'Antonio JM, Ma C, Monzon FA, et al. Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate 2008;68:698-714. [Crossref] [PubMed]
- Chandran UR, Ma C, Dhir R, et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007;7:64. [Crossref] [PubMed]
- Yu YP, Landsittel D, Jing L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 2004;22:2790-9. [Crossref] [PubMed]
- Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005;8:393-406. [Crossref] [PubMed]
- Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A 2004;101:811-6. [Crossref] [PubMed]
- Malhotra S, Lapointe J, Salari K, et al. A tri-marker proliferation index predicts biochemical recurrence after surgery for prostate cancer. PLoS One 2011;6:e20293. [Crossref] [PubMed]
- Tomlins SA, Mehra R, Rhodes DR, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 2007;39:41-51. [Crossref] [PubMed]
- Shibue T, Brooks MW, Weinberg RA. An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell 2013;24:481-98. [Crossref] [PubMed]
- Troussard AA, Mawji NM, Ong C, et al. Conditional knock-out of integrin-linked kinase demonstrates an essential role in protein kinase B/Akt activation. J Biol Chem 2003;278:22374-8. [Crossref] [PubMed]
- Li P, Yang R, Gao WQ. Contributions of epithelial-mesenchymal transition and cancer stem cells to the development of castration resistance of prostate cancer. Mol Cancer 2014;13:55. [Crossref] [PubMed]
- Wang XS, Shankar S, Dhanasekaran SM, et al. Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov 2011;1:35-43. [Crossref] [PubMed]
- Noguchi S, Yasui Y, Iwasaki J, et al. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett 2013;328:353-61. [Crossref] [PubMed]
- Tang Q, Zhao S, Wu J, et al. Inhibition of integrin-linked kinase expression by emodin through crosstalk of AMPKalpha and ERK1/2 signaling and reciprocal interplay of Sp1 and c-Jun. Cell Signal 2015;27:1469-77. [Crossref] [PubMed]
- Clarke DM, Brown MC, LaLonde DP, et al. Phosphorylation of actopaxin regulates cell spreading and migration. J Cell Biol 2004;166:901-12. [Crossref] [PubMed]
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Sun Y, Ding Y, Guo C, Liu C, Ma P, Ma S, et al. alpha-Parvin promotes breast cancer progression and metastasis through interaction with G3BP2 and regulation of TWIST1 signaling. Oncogene 2019;38:4856-74. [Crossref] [PubMed]
- Zhang Y, Chen K, Tu Y, et al. Distinct roles of two structurally closely related focal adhesion proteins, alpha-parvins and beta-parvins, in regulation of cell morphology and survival. J Biol Chem 2004;279:41695-705. [Crossref] [PubMed]
- Cortez V, Nair BC, Chakravarty D, et al. Integrin-linked kinase 1: role in hormonal cancer progression. Front Biosci (Schol Ed) 2011;3:788-96. [PubMed]
- Hu MB, Hu JM, Jiang LR, et al. Differential expressions of integrin-linked kinase, beta-parvin and cofilin 1 in high-fat diet induced prostate cancer progression in a transgenic mouse model. Oncol Lett 2018;16:4945-52. [PubMed]
- Becker-Santos DD, Guo Y, Ghaffari M, et al. Integrin-linked kinase as a target for ERG-mediated invasive properties in prostate cancer models. Carcinogenesis 2012;33:2558-67. [Crossref] [PubMed]
- Persad S, Attwell S, Gray V, et al. Inhibition of integrin-linked kinase (ILK) suppresses activation of protein kinase B/Akt and induces cell cycle arrest and apoptosis of PTEN-mutant prostate cancer cells. Proc Natl Acad Sci U S A 2000;97:3207-12. [Crossref] [PubMed]
- Vaynberg J, Fukuda K, Lu F, et al. Non-catalytic signaling by pseudokinase ILK for regulating cell adhesion. Nat Commun 2018;9:4465. [Crossref] [PubMed]
- Velazquez-Torres G, Shoshan E, Ivan C, et al. A-to-I miR-378a-3p editing can prevent melanoma progression via regulation of PARVA expression. Nat Commun 2018;9:461. [Crossref] [PubMed]
- Bluemn EG, Coleman IM, Lucas JM, et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017;32:474-489.e6. [Crossref] [PubMed]
- Gao H, Ouyang X, Banach-Petrosky WA, et al. Combinatorial activities of Akt and B-Raf/Erk signaling in a mouse model of androgen-independent prostate cancer. Proc Natl Acad Sci U S A 2006;103:14477-82. [Crossref] [PubMed]
- Wang W, Liu Q, Zhang Y, et al. Involvement of ILK/ERK1/2 and ILK/p38 pathways in mediating the enhanced osteoblast differentiation by micro/nanotopography. Acta Biomater 2014;10:3705-15. [Crossref] [PubMed]
- Persad S, Attwell S, Gray V, et al. Regulation of protein kinase B/Akt-serine 473 phosphorylation by integrin-linked kinase: critical roles for kinase activity and amino acids arginine 211 and serine 343. J Biol Chem 2001;276:27462-9. [Crossref] [PubMed]
- Attwell S, Mills J, Troussard A, et al. Integration of cell attachment, cytoskeletal localization, and signaling by integrin-linked kinase (ILK), CH-ILKBP, and the tumor suppressor PTEN. Mol Biol Cell 2003;14:4813-25. [Crossref] [PubMed]
- Pereira JA, Benninger Y, Baumann R, et al. Integrin-linked kinase is required for radial sorting of axons and Schwann cell remyelination in the peripheral nervous system. J Cell Biol 2009;185:147-61. [Crossref] [PubMed]
(English Language Editor: J. Gray)


