
Effect of myeloid ecotropic viral integration site (MEIS) family genes on tumor microenvironment remodeling and its potential therapeutic effect
Introduction
The homeobox genes were discovered in Drosophila melanogaster and were so named owing to their high spatio-temporal specificity (1). They are present in almost all eukaryotic cells (2) and encode proteins that bind to the promoter region of specific genes and act as transcription factors. Therefore, the homeobox genes are critical in the development of the organism (3). Studies have shown that several homeobox genes are abnormally expressed in cancer and contribute to its progression.
The homeobox genes are classified into two types—HOX genes and non-HOX genes or HOX branching genes. The myeloid ecotropic viral integration site (MEIS) family (MEIS1, MEIS2, and MEIS3) belongs to the HOX branching genes, and their abnormal expression is closely related to the development of many types of cancers (4). As the MEIS proteins are highly similar (MEIS1 and MEIS2 are more than 85% identical in mice and humans), it is challenging to distinguish between them (5). With this in mind, in this study, we use MEIS to refer to all MEIS family members and use the individual gene names only if the MEIS member has specific functions.
MEIS1 is an oncogene whose frequent integration and overexpression with HoxA9 can lead to invasive leukemia (acute lymphoblastic leukemia) in mice and humans. In addition, MEIS1 is also expressed in many cancers, such as prostate, pancreatic, colorectal, and breast cancer, and other solid tumors (6-8). Similarly, it has been reported that MEIS2 is abnormally expressed in prostate cancer, ovarian cancer, and neuroblastomas (6,9,10), suggesting that MEIS2 is closely related to the occurrence and development of cancers. Halder et al. reported that MEIS2 contributes to the development of lung cancer by inhibiting the expression of transforming growth factor β type II receptor (11). In addition, Vegi et al. reported that MEIS2 participates in the development of leukemia by promoting the proliferation of myeloid cells and inhibiting their differentiation, suggesting that MEIS2 may be a target for leukemia therapy (12).
Compared to MEIS1 and MEIS2, there are a limited number of studies on the role of MEIS3 in cancer development. Lu et al. found that MEIS3 promotes gene transcription by binding to the cg02515217 CpG site in the MIR-21 (carcinogenic microRNA) promoter, resulting in a significant increase in its expression in a variety of cancers (13). In addition, Liu et al. also found that the expression of MEIS3 is related to the survival of ovarian cancer cells (14).
Abnormal expression of the MEIS family members has been reported in many cancers. To date, there has been no systematic study of this gene family in different human cancers, and each gene has only been studied in a few cancers, most of which were carried out using cell lines and/or animal models. Therefore, in an effort to understand the global role of MEIS in the development and progression of cancer, we used 33 primary tumor data obtained from The Cancer Genome Atlas (TCGA) database and determined the differential expression of this gene family in different cancers and their relationship with the overall survival rate of patients. In addition, we also explored the correlation between the expression level of different MEIS genes and the tumor microenvironment (TME) and drug sensitivity. Our results shed light on the role of the MEIS family in different cancers, especially with respect to immune response, TME, and drug resistance, providing insights to strategies for developing personalized cancer drugs.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1163).
Methods
Data sources
TCGA pan-cancer data was downloaded from the Xena browser (https://xenabrowser.net/datapages/). The data included RNA-seq data, clinical data, stemness scores based on mRNA (RNAss) and DNA methylation (DNAss), and immune subtypes of 33 cancer types—ACC, BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC, UCS, and UVM (see Table S1 for details). A survival analysis of all samples was carried out to investigate the relationship between gene expression (as a continuous variable) of each MEIS family member and overall survival.
Because the data from TCGA are publicly available and open-access, this study do not require approval by the local ethics committees. This study followed TCGA data access policies and publication guidelines.
Expression level, difference analysis, and correlation analysis of the MEIS family genes in pan-cancer
After obtaining the data, we first visualized the expression and distribution of the MEIS family genes in 33 types of cancers using a boxplot graph. Next, based on log2 (fold change), we drew heat maps of 18 tumor types with more than five adjacent normal samples to show the difference in MEIS gene expression between primary tumors and adjacent normal tissues. Finally, we used Spearman’s correlation test to calculate the gene expression correlation of three MEIS family members in 33 cancer types and visualized them.
The relationship between the MEIS family and survival rate of patients
Patients with 33 different cancers were divided into high and low MEIS expression groups according to the median expression of three members of the MEIS family, and a Kaplan-Meier survival curve was plotted to compare the survival differences between the two groups. In addition, we also used a univariate Cox proportional hazard regression model to detect the relationship between gene expression and overall survival.
Correlation analysis of immune infiltration and TME
The analysis of variance was used to test the correlation between MEIS expression and immune infiltrating subtypes in all cancer types to further understand the relationship between MEIS family members and immune components in tumors. The estimation of stromal-immune cells and immune cells in malignant tumors using expression data (ESTIMATE) algorithm is a method that uses gene expression information to infer the ratio of stromal cells and immune cells in tumor samples. We used the ESTIMATE algorithm to score the stroma and immunity of each sample. We then used the Spearman correlation test to analyze the infiltration level of immune and stromal cells in different tumors (15).
Correlation analysis of tumor stem cells and drug sensitivity analysis
To explore the characteristics of various tumor stem cells, we extracted the transcriptome and epigenetic methylation data of TCGA tumor samples. We then performed Spearman correlation tests to detect the correlation between tumor stem cell characteristics and MEIS expression. In addition, we downloaded the data of different cancer cell lines from the NCI-60 database (https://discover.nci.nih.gov/cellminer/) and used the Pearson correlation test to explore the relationship between MEIS expression and drug sensitivity. We included 263 drugs approved by the FDA or drugs in clinical trials for correlation analysis.
Relationship between MEIS family and clear cell renal cell carcinoma (ccRCC)
In order to explore the correlation between MEIS expression and a certain type of cancer that we are concerned about (ccRCC in this study), we analyzed and visualized the correlation between MEIS family and immune subtypes, clinical characteristics, TME, and stem cells of a single tumor. In addition, to further explore the content of immune cells in a certain type of cancer, we used the CIBERSORT calculation method to analyze the proportion of tumor-infiltrating immune subsets. At the same time, quality filtering was performed, and only 539 tumor samples with P<0.05 were analyzed.
Statistical analysis
Statistical analyses were performed using the SPSS software v. 22.0 (Chicago, IL, USA) and the R package v. 3.6.1 (https://www.r-project.org/). The linear mixed-effects model was used to analyze the differences in gene expression between tumor and normal tissues. Univariate and multivariate Cox regression analyses or the Log-ranch test were used to investigate the relationship between gene expression and the patients’ overall survival. The association between gene expression and the stemness, stromal, immune, and estimate scores, as well as drug sensitivity, was given via the calculation of Spearman’s or Pearson’s correlation coefficients. In addition, linear regression was used to investigate the relationship between gene expression and the patients’ clinical characteristics, immune components, and ccRCC subtypes. Statistical significance is defined as P<0.05.
Ethical statement
All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and no ethical approval was required because the data we used were obtained from public databases. Because of the retrospective nature of the research, the requirement for informed consent was waived.
Results
Expression of MEIS family in pan-cancer
To explore the heterogeneity of the MEIS family, we detected the expression level of MEIS family members in all 33 cancer types in TCGA pan-cancer data. We observed that all three MEIS family genes were overexpressed in cancers (Figure 1A). There was significant heterogeneity in the expression levels of each member of the MEIS family. In some tumor types, the level of expression of a particular gene was very high, whereas in other tumor types, the gene expression level was significantly downregulated or had no significant change (Figure 1B,C,D). These findings indicate that each gene needs to be studied as a separate entity in the future.
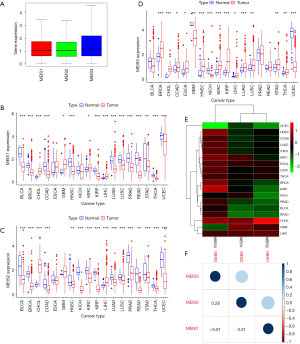
All MEIS family genes showed significant differential expression in different types of cancer, but the expression of each gene changed in different directions in different cancers. For example, MEIS3 was upregulated in most tumors, whereas MEIS2 was downregulated (Figure 1E). In addition, through the Spearman correlation test, we found that the expression levels of members of the MEIS family were positively correlated in different types of cancer (Figure 1F).
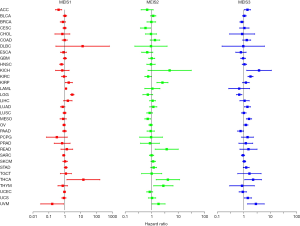
Relationship between MEIS expression and overall survival rate of patients
To correlate and ultimately predict which MEIS family members promote or inhibit tumorigenesis in which cancer types, we used 33 types of primary tumors to study the relationship between MEIS expression and overall survival. We found that changes in MEIS expression usually affect overall survival. As shown in Figure 2, whether a member of the MEIS family is a risk factor or a protective factor varies depending on the type of cancer.
The MEIS family is associated with immune response and TME
To understand the relationship between MEIS family members and immune components, we tested the correlation between MEIS and immune infiltration in tumors. Six different types of immune infiltration have been found in human tumors, corresponding to tumor promotion and inhibition. They are C1 (wound healing), C2 (INF-r dominant), C3 (inflammation), C4 (lymphopenia dominant), C5 (immunologically quiet), and C6 (TGFβ dominant) (16). We correlated the immune infiltration in TCGA pan-cancer data with the expression levels of MEIS family members and visualized it (Figure 3A). The high expression of the MEIS family is related to the upregulation of C5. In particular, high expression of MEIS2 and MEIS3 is associated with immune silencing, suggesting that the role of these genes in promoting or inhibiting cancer may be related to their immune effects.
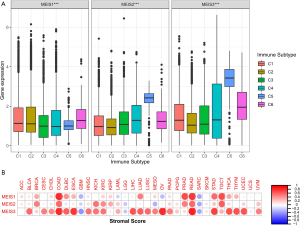
In addition, we used the ESTIMATE algorithm to study the relationship between the expression level of MEIS and invasive stromal cells in tumors (Figure 3B). We found that the expression levels of the MEIS family members are very different from the StromalScore of different cancer types. Among them, the correlation between MEIS3 and StromalScore of various cancer types was the highest.
MEIS family is associated with tumor stem cells and chemosensitivity
In the process of cancer development, tumor cells can gradually lose their differentiation phenotype and obtain progenitor and stem cell-like characteristics. The characteristics of tumor stem cells can be measured via RNA stemness score (RNAss) based on mRNA expression and DNA stemness (DNAss) depending on the DNA methylation pattern (17). We found that members of the MEIS family were associated with RNAss and DNAss to varying degrees in different types of cancer (Figure 4A,B). It is worth noting that almost all members of the MEIS family are negatively correlated with RNAss (r<0). In addition, in STAD and TGCT, all MEIS genes were negatively correlated with DNAss and RNAss, whereas in THYM, genes were positively correlated with DNAss and negatively correlated with RNAss.
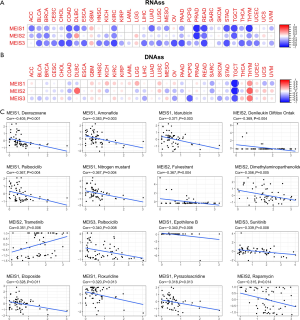
At the same time, we explored the relationship between MEIS expression and drug sensitivity. Here, we sort the first 16 most relevant ones according to their P values (Figure 4C). Further, with the increase in the expression of MEIS family, especially the higher the expression of MEIS1 and MEIS2, the less sensitive the cells are to chemotherapeutic drugs.
Role of the MEIS family in ccRCC
A few previous studies have explored the role of the MEIS family in renal tumors (13,18,19). However, the sample sizes of the previous studies were small, and most of them were carried out in cell lines or animal models. Therefore, we used data from ccRCC in TCGA to explore the relationship between ccRCC and the MEIS family. Figure 1 shows that there is a significant difference in the expression of all MEIS genes between ccRCC tumors and adjacent normal cells (all P<0.05).
In addition, we further determined the correlation between the MEIS family and various immune subtypes in the ccRCC TME. The results showed that there were significant differences among different immune subtypes of the MEIS family (Figure 5A). It is worth noting that the relationship between MEIS gene expression and immune subtypes in ccRCC contradicts that observed in the pan-cancer data, that is, the low expression of C5 in MEIS2 and MEIS3.
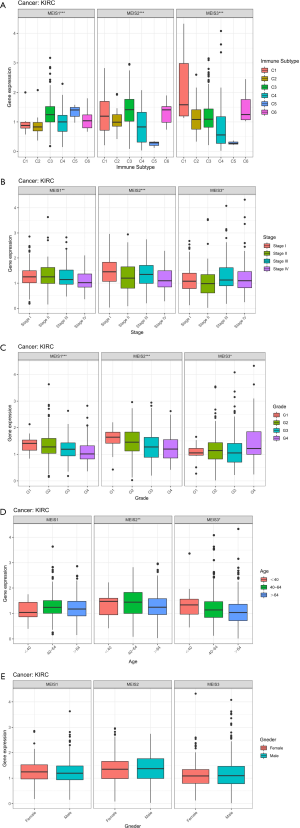
We further analyzed the relationship between the clinicopathological parameters of ccRCC and the expression of MEIS family. The results showed that the expression of MEIS genes decreased with an increase in the clinical stage, pathological grade, and age (Figure 5B,C,D). This suggests that the overexpression of MEIS genes may be related to better clinical traits. In addition, the analysis showed that the expression of MEIS genes was not related to sex (Figure 5E).
We then determined the correlation between the expression of MEIS genes and TME and stem cell index in patients with ccRCC. We found that the expression of MEIS genes was positively correlated with StromalScore (Figure 6A). The expression of MEIS2 and MEIS3 was also significantly correlated with tumor purity (ESTIMATEscore) (P<0.001).
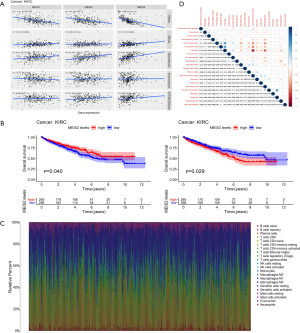
Survival analysis of ccRCC shows that an increase in MEIS3 expression tends to decrease the survival rate, whereas increases in MEIS1 and MEIS2 expression levels tends to improve the survival rate. However, only MEIS2 and MEIS3 showed significant associations (Figure 6B).
At the same time, we found that members of the MEIS family were negatively correlated with RNAss and that a decrease in tumor stemness was usually associated with better survival. This contradicts the fact that the overexpression of MEIS3 reduces the survival rate of patients, suggesting that MEIS3 might modulate the overall survival through other mechanisms.
In addition, by analyzing the proportion of tumor-infiltrating immune subsets using the CIBERSORT algorithm, we constructed 22 types of immune cell maps of ccRCC samples and analyzed the correlation between immune cells (Figure 6C,D). The results showed that the two types of immune cells with the strongest positive correlation were CD8 T cells and follicular helper T cells (r=0.55), and the two types of immune cells with the strongest negative correlation were CD8 T cells and CD4 memory resting T cells (r=−0.71).
Discussion
The MEIS family is a member of the triple amino acid loop extension (TALE) transcription factors, which contain a homologous domain and play an important role in cell growth and differentiation during vertebrate embryogenesis. Previous studies have confirmed that it plays a key role in tumor growth, metastasis, and diffusion, including participating in controlling tumor cell viability, inhibiting cell proliferation, and inducing cell cycle arrest. However, the relationship between the MEIS family and tumor compartment and microenvironment has not been reported.
In our study, we systematically analyzed the pan-cancer expression of the MEIS family for the first time. We found that there was significant heterogeneity in MEIS gene expression between different tumor types and within each tumor type. For example, MEIS3 is upregulated in most tumors, whereas MEIS2 is downregulated. In addition, we found that their expression was different in different pathological types of the same cancer, such as KICH, KIRC, and KIRP. A similar conclusion was also reported by Schulte et al. by analyzing studies on MEIS in different cancers. It was found that the expression of the same MEIS gene may be increased in some cancer types and downregulated in other cancer types (5). At the same time, literature analysis revealed that even in the same tumor, the same MEIS gene could play the role of proto-oncogene and tumor suppressor gene (20-22). We further tested the relationship between MEIS expression levels and overall survival of patients, and the results showed that changes in MEIS expression were usually related to overall survival of patients, and the direction of the association was dependent on the type of cancer. We speculate that the reason for this phenomenon is that the activity of MEIS proteins may depend on the cell type and that the function of these proteins is determined by the cellular environment in which these proteins are located. This suggests that the challenge for cancer researchers may be to evaluate the function of MEIS proteins for different cell and tumor types, even for different subtypes of the same cancer.
TME is the environment where tumor cells survive, and mainly include tumor cells, many non-tumor cells (such as T cells, tumor-related fibroblasts, and dendritic cells), tumor extracellular matrix, and blood vessels (23,24). The relationship between the tumor and its microenvironment is like that between the “seed” and “soil”. The microenvironment provides nutrition and conditions for tumor growth. In the process of tumor cell growth and migration, non-tumor cells (mainly stromal cells and immune cells) can enter the tumor tissue; this phenomenon is called infiltration. The infiltration of stromal cells and immune cells is closely related to the clinical outcome (25,26), and of great value in tumor diagnosis and prognosis, and may be used as a drug target to improve the survival rate of patients.
TME plays a key role in the initiation and progression of cancers. It is of great significance to explore the potential therapeutic targets of TME remodeling and promote the transformation of TME from a tumor-friendly to tumor suppressor type (25). Our study found that all MEIS family members were associated with immune infiltration subtypes, stromal cells, and immune cell infiltration levels in the TME. Specifically, the high expression of the MEIS family is associated with the upregulation of C5. Previous studies by Lehrer et al. have reported that high levels of C5 (immune silencing) will weaken the efficacy of immune checkpoint inhibitors (27). It is suggested that for patients with advanced cancer, interfering with the expression of MEIS while using immune checkpoint inhibitors may benefit the patients. However, this idea still needs to be confirmed by further experiments. Stromal cells have been shown to play an important role in tumor growth, disease progression, and drug resistance. In addition, infiltrating immune cells are also related to tumor invasion and metastasis (28,29). These results suggest that the MEIS family may be a therapeutic target or help predict the efficacy of immune checkpoint modulators in cancer patients.
Analysis of MEIS family expression, tumor Stemness score, and drug sensitivity, showed that an increase in MEIS family expression, weakens the stemness characteristics, albeit making the cancer less sensitive to chemotherapeutic drugs. This seems to be contradictory, but it also explains to some extent, the heterogeneity of the impact of the MEIS family on the survival of patients with different cancers. Although the results suggest that MEIS may be a potential key target for treatment and prognosis, all findings need to be further verified by relevant experiments.
MEIS1 has been widely studied; Zhu et al. reported that MEIS1 might play an anticancer role in ccRCC (18). In this study, we found that the expression of MEIS1 in ccRCC tissue or cell line decreased, and the overexpression of MEIS1 inhibited the proliferation of ccRCC cells. This is somewhat different from the results of our study. The analysis of TCGA pan-cancer data shows that the expression of MEIS1 in ccRCC tissue is increased, although the increased expression of MEIS1 tends to improve the survival rate of patients; however, no significant difference was observed in the survival. We speculate that the reason for the above phenomenon is that the function of MEIS1 in cancer cells is affected by many factors, and its complex role in proliferation may largely depend on the TME.
In recent years, significant progress has been made in immunotherapy. Immune checkpoint modulators have become an important part of modern tumor therapy and have been approved for the treatment of various cancers, including ccRCC (30). Previous studies have reported that the increased density of CD8+ T cells in TME is associated with a good prognosis of ccRCC (31,32). However, several immune cells have the opposite effect. For example, infiltrating CD4+ T cells in TME can promote the proliferation of RCC cells by regulating TGF β 1/YBX1/HIF2 α signaling (33). In addition, regulatory T cells (Tregs) can also inhibit tumor immune response by releasing immunosuppressive cytokines (34). We visualized immune cell infiltration in patients with ccRCC using the CIBERSORT algorithm, and further analyzed the correlation between immune cells. This can help us to understand the composition and interaction of various immune cells more intuitively in the TME of such patients, which may identify new therapeutic targets and ideas for immunotherapy in patients with ccRCC.
At the same time, as mentioned above, almost all members of the MEIS family are negatively correlated with RNAss, suggesting that MEIS may be related to the sensitivity or resistance of cancer cells to chemotherapy. In addition, we found that the increased expression of MEIS genes, especially the expression of MEIS1 and MEIS2, led to an increase in cell resistance to chemotherapeutic drugs. Of course, different members of the MEIS family are also associated with increased drug sensitivity of some drugs. For example, the increased expression of MEIS2 is related to the increased sensitivity of cancer cells to trametinib. Trametinib was approved by the FDA in 2013 for the treatment of unresectable or metastatic melanoma with BRAF V600E or V600K gene mutations (35). It has also been reported to have satisfactory efficacy in lung cancer, colorectal tumors, gliomas, and other malignant tumors (36-38). These data suggest that MEIS may play a role in the sensitivity or drug resistance of tumor cells to drug therapy, and can be used as a therapeutic target to overcome drug resistance or help to increase drug sensitivity.
In this study, we discussed the differences in MEIS family expression in different tumors and their possible biological functions. We also focused on exploring the immune and TME, clinicopathological features, and tumor stem cell correlation of this gene family in patients with ccRCC. Nevertheless, this study has some limitations. First, all patient data are derived from public retrospective study data, so the possibility of selection bias cannot be ruled out. Second, because there are too many cancer types included, the degree of integrity of patient information varies among different cancer groups, and there is no normal sample data for several cancer types, these cancer types cannot be further analyzed. Third, as almost all patients in the database are Caucasian, the application of the conclusions in other populations needs to be further explored. Finally, we did not conduct experimental studies to explore the function of the MEIS family in ccRCC. Therefore, future studies need to be carried out in vitro and in vivo to explore the detailed mechanism between the expression of MEIS family and the occurrence and development of ccRCC, and to prove the conclusions of this study, in order to provide a direction for exploring the mechanism of this gene family in other cancers.
Conclusions
TCGA pan-cancer dataset will continue to be used as a resource for the exploration of human cancer. Using this data, we explored and discussed the pan-cancer expression and molecular characteristics of the MEIS gene family and its effects on immunity and TME. Our results show that the MEIS family plays different roles in different types of cancer, and we also found that their expression is different in different pathological types of the same cancer. This study will help to reveal the role of this gene family in tumorigenesis and development, especially in the immune response, TME, and drug resistance. Therefore, it provides ideas for carrying out relevant laboratory research to confirm the pertinent ideas and is also essential for the development of personalized cancer treatment drugs.
Acknowledgments
Funding: This work was supported by the National Key R&D Program of China (grant number 2018YFC2002202).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1163
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1163). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and no ethical approval was required because the data we used were obtained from public databases. Because of the retrospective nature of the research, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gorski DH, Walsh K. The role of homeobox genes in vascular remodeling and angiogenesis. Circ Res 2000;87:865-72. [Crossref] [PubMed]
- Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer 2005;41:2428-37. [Crossref] [PubMed]
- Tucker SC, Wisdom R. Site-specific heterodimerization by paired class homeodomain proteins mediates selective transcriptional responses. J Biol Chem 1999;274:32325-32. [Crossref] [PubMed]
- Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med 2014;92:811-23. [Crossref] [PubMed]
- Schulte D, Geerts D. MEIS transcription factors in development and disease. Development 2019;146:dev174706. [Crossref] [PubMed]
- Chen JL, Li J, Kiriluk KJ, et al. Deregulation of a Hox protein regulatory network spanning prostate cancer initiation and progression. Clin Cancer Res 2012;18:4291-302. [Crossref] [PubMed]
- Tomoeda M, Yuki M, Kubo C, et al. Role of Meis1 in mitochondrial gene transcription of pancreatic cancer cells. Biochem Biophys Res Commun 2011;410:798-802. [Crossref] [PubMed]
- Crist RC, Roth J, Waldman SA, et al. A conserved tissue-specific homeodomain-less isoform of MEIS1 is downregulated in colorectal cancer. PLoS One 2011;6:e23665. [Crossref] [PubMed]
- Crijns AP, de Graeff P, Geerts D, et al. MEIS and PBX homeobox proteins in ovarian cancer. Eur J Cancer 2007;43:2495-2505. [Crossref] [PubMed]
- Zha Y, Xia Y, Ding J, et al. MEIS2 is essential for neuroblastoma cell survival and proliferation by transcriptional control of M-phase progression. Cell Death Dis 2014;5:e1417. [Crossref] [PubMed]
- Halder SK, Cho YJ, Datta A, et al. Elucidating the mechanism of regulation of transforming growth factor β Type II receptor expression in human lung cancer cell lines. Neoplasia 2011;13:912-22. [Crossref] [PubMed]
- Vegi NM, Klappacher J, Oswald F, et al. MEIS2 Is an Oncogenic Partner in AML1-ETO-Positive AML. Cell Rep 2016;16:498-507. [Crossref] [PubMed]
- Lu J, Tan T, Zhu L, et al. Hypomethylation Causes MIR21 Overexpression in Tumors. Mol Ther Oncolytics 2020;18:47-57. [Crossref] [PubMed]
- Liu J, Wang Y, Birnbaum MJ, et al. Three-amino-acid-loop-extension homeodomain factor Meis3 regulates cell survival via PDK1. Proc Natl Acad Sci 2010;107:20494-99. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Tamborero D, Rubio-Perez C, Muiños F, et al. A pan-cancer landscape of interactions between solid tumors and infiltrating immune cell populations. Clin Cancer Res 2018;24:3717-28. [Crossref] [PubMed]
- Malta TM, Sokolov A, Gentles AJ, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 2018;173:338-354.e15. [Crossref] [PubMed]
- Zhu J, Cui L, Xu A, et al. MEIS1 inhibits clear cell renal cell carcinoma cells proliferation and in vitro invasion or migration. BMC Cancer 2017;17:176. [Crossref] [PubMed]
- Dekel B, Metsuyanim S, Schmidt-Ott K M, et al. Multiple imprinted and stemness genes provide a link between normal and tumor progenitor cells of the developing human kidney. Cancer Res 2006;66:6040-49. [Crossref] [PubMed]
- Bhanvadia RR, Vanopstall C, Brechka H, et al. MEIS1 and MEIS2 expression and prostate cancer progression: a role for HOXB13 binding partners in metastatic disease. Clin Cancer Res 2018;24:3668-80. [Crossref] [PubMed]
- Johng D, Torga G, Ewing CM, et al. HOXB13 interaction with MEIS1 modifies proliferation and gene expression in prostate cancer. Prostate 2019;79:414-24. [Crossref] [PubMed]
- Jeong JH, Park SJ, Dickinson SI, et al. A constitutive intrinsic inflammatory signaling circuit composed of miR-196b, Meis2, PPP3CC, and p65 drives prostate cancer castration resistance. Mol Cell 2017;65:154-67. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- Wood SL, Pernemalm M, Crosbie PA, et al. The role of the tumor microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev 2014;40:558-66. [Crossref] [PubMed]
- Bi KW, Wei XG, Qin XX, et al. BTK has potential to be a prognostic factor for lung adenocarcinoma and an indicator for tumor microenvironment remodeling: a study based on TCGA data mining. Front Oncol 2020;10:424. [Crossref] [PubMed]
- Wang H, Wu X, Chen Y. Stromal-immune score-based gene signature: a prognosis stratification tool in gastric cancer. Front Oncol 2019;9:1212. [Crossref] [PubMed]
- Lehrer S, Rheinstein P H. Expression of the vesicular monoamine transporter gene solute carrier family 18 member 1 (SLC18A1) in lung cancer. Cancer Genomics Proteomics 2018;15:387-93. [Crossref] [PubMed]
- Kobayashi H, Enomoto A, Woods S L, et al. Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol 2019;16:282-95. [Crossref] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. [Crossref] [PubMed]
- Sadreddini S, Baradaran B, Aghebati-Maleki A, et al. Immune checkpoint blockade opens a new way to cancer immunotherapy. J Cell Physiol 2019;234:8541-49. [Crossref] [PubMed]
- Youngblood B, Hale JS, Kissick HT, et al. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 2017;552:404-9. [Crossref] [PubMed]
- Yao J, Xi W, Zhu Y, et al. Checkpoint molecule PD-1-assisted CD8(+) T lymphocyte count in tumor microenvironment predicts overall survival of patients with metastatic renal cell carcinoma treated with tyrosine kinase inhibitors. Cancer Manag Res 2018;10:3419-31. [Crossref] [PubMed]
- Wang Y, Wang Y, Xu L, et al. CD4 + T cells promote renal cell carcinoma proliferation via modulating YBX1. Exp Cell Res 2018;363:95-101. [Crossref] [PubMed]
- Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol 2016;16:599-611. [Crossref] [PubMed]
- Robert C, Grob JJ, Stroyakovskiy D, et al. Five- year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med 2019;381:626-36. [Crossref] [PubMed]
- Kondyli M, Larouche V, Saint-Martin C, et al. Trametinib for progressive pediatric low-grade gliomas. J Neurooncol 2018;140:435-44. [Crossref] [PubMed]
- Kelly RJ. Dabrafenib and trametinib for the treatment of non-small cell lung cancer. Expert Rev Anticancer Ther 2018;18:1063-8. [Crossref] [PubMed]
- Lin L, Ding D, Xiao X, et al. Trametinib potentiates TRAIL-induced apoptosis via FBW7-dependent Mcl-1 degradation in colorectal cancer cells. J Cell Mol Med 2020;24:6822-32. [Crossref] [PubMed]


