A potential panel of five mRNAs in urinary extracellular vesicles for the detection of bladder cancer
Introduction
As the 10th leading cancer worldwide, bladder cancer is estimated to have 549,000 new cases and be responsible for 200,000 deaths in 2018 (1). From statistics, the incidence and mortality rates of bladder cancer in men are respectively 9.6 and 3.2 per 100,000, nearly three times higher than women. There is also evidence showing that bladder cancer is more common in developed countries, such as Europe and Northern America. In stark contrast to the incidence rate, bladder cancer has the highest lifetime treatment cost per patient among all cancers (2). Early diagnosis and treatment is still the most effective way to improve prognosis and reduce medical expenditure for treatment.
Till now, cystoscopy remains the primary method for the diagnosis and recurrence monitoring of bladder cancer. The effectiveness of cystoscopy is operator dependent, thus the actual sensitivity and specificity may vary due to operator-induced factor (3). Moreover, as an invasive inspection, possible adverse effects include infection, whilst dysuria, frequency and visible hematuria (4,5). As supplementary measures, a number of urinary based non-invasive tests had been developed to diagnose bladder cancers. For example, urine cytology is an effective tool for the detection of high-grade and high-stage bladder cancer, but ineffective for low-grade malignancy (6,7). UroVysion, a fluorescence in situ hybridization (FISH) probe developed in 2000, allows the detection of bladder cancer by examining the chromosomal aberrations of exfoliated urothelial cells. This test possesses a relatively high specificity, but still has difficulty in the diagnosis of low-grade bladder cancers (8). Additional assays, such as NMP22 and BTA test also suffered from false positive caused by other diseases of urinary system.
Extracellular vesicles (EVs), known as cell-derived membranous structures carrying a variety of biomolecules, have emerged as a novel analyte in liquid biopsy during the past decade (9-11). Recently, a growing number of evidence suggests the important role of EVs in bladder cancer detection. For example, using a double-filtration microfluidic device, Wang and his group quantitatively analyzed the concentration of urinary EVs, which is notably elevated in bladder cancer patients. In addition to the absolute quantity, different signatures of EVs associated biomolecules between bladder cancer patients and healthy controls had also been reported (12-14). Particularly, Murakami et al. had introduced a method to identify mRNA biomarkers by urinary EV RNA-seq analysis (15). In this work, urinary EVs isolated from 12 samples, including non-muscle invasive bladder cancer (N=3), advanced urothelial cancer (N=3), no residual tumor after TURBT (N=2), and healthy or disease controls (N=4), were subjected to RNA sequencing. Considering that the presence of EVs with other origins (non-bladder cancer cells) is inevitable in urine, the sequencing data is likely to reflect the signature of a mixture of heterogeneous vesicles. With a very limited sample size, it is hard to guarantee the differentially expressed genes are indeed caused by bladder cancer instead of other unidentified factors.
Herein, we introduced a method to develop a multivariable prediction model based on potential RNA markers identified from tissue RNA Sequencing data. Unlike the sequencing of a few EVs samples, tissue sequencing is well established, has stringent quality control, and more importantly has a large amount of available data in public database. In light of these, we first examined the expression level of identified candidate markers in urinary EVs collected from a training cohort (n=368, 126 bladder cancer and 242 negative controls). Logistic regression model was established based on the qPCR results. The optimal diagnostic model (ExoPanel) developed in this process was further validated by a validation cohort (n=155, 56 bladder cancer and 99 negative controls). The diagnostic performance of the ExoPanel was promising and in well accordance between training cohort and validation cohort. We present the following article in accordance with the TRIPOD reporting checklist (available at: http://dx.doi.org/10.21037/tau-20-1057).
Methods
Study population
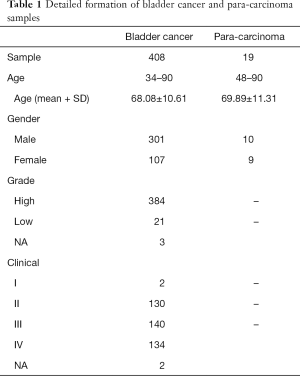
From January 2017 to September 2019, the study recruited 182 bladder cancer patients, 152 healthy control, 64 disease controls, and 125 non-recurrent bladder cancer patients after TURBT. The clinical outcome of bladder cancer patients and disease control were diagnosed by cystoscopy. Patients with non-urothelial bladder cancer, infectious virus positive patients, or patients receiving chemotherapy within 2 months were excluded from this study. All participants were divided into either bladder cancer group or negative control group. Then, participants in each group were randomly divided into training cohort (n=368, 126 bladder cancer and 242 negative controls) or validation cohort (n=155, 56 bladder cancer and 99 negative controls) at a ratio of 7:3. All procedures performed in this study were in accordance with the Declaration of Helsinki (revised in 2013). This study was approved by the ethics committee of Chinese PLA General Hospital (C2017-037-01). Informed consents are obtained from all participants.
Isolation of urinary EVs
For each individual, 80 mL first-void mid-stream urine samples is collected, stored on ice and immediately delivered to the lab. For patients about to undergo cystoscopy test, the urine sample was collected one day in advance. Urinary EVs are isolated according to a modified protocol as reported (16). Briefly, the sediment of urine is removed by centrifugation at 2,000×g for 20 min. 30 mL supernatant is then transferred to a new tube and mixed PEG 6000 40% wt at 4:1 ratio by volume. After incubation at 4 °C overnight, the solution is centrifuged at 4,500 ×g for 20 min. The supernatant is discarded, and the pellet is collected for either EVs characterization or RNA extraction.
Characterization of urinary EVs
Transmission electron microscopy (TEM)
Urinary EVs pellet is re-suspended in PBS. A total of 20 µL EVs solution is added on top of a copper mesh with a pipette. After 10 min incubation at room temperature, the solution is drained, and washed with sterile distilled water. The sample is then contrasted by uranyl-oxalate solution for 1 min, dried for 2 min under incandescent light, and finally imaged under transmission electron microscope (H-7650, HITACHI, Japan).
Nano-particle tracking analysis (NTA)
Urinary EVs solution is first diluted to a desire concentration between 1×107/mL to 1×109/mL. The solution is then loaded into the sample cell of a ZetaView PMX 110 (Particle Metrix, Germany). A 60-seconds video is recorded with a frame rate of 30 frames per second. The size and concentration of EVs is calculated by NTA software (ZetaView 8.02.28).
Western blot analysis (WB)
The EVs solution is mixed with sodium dodecyl sulfonate (SDS) buffer and subjected to western blot analysis (10% SDS-polyacrylamide gel electrophoresis; 50 µg protein/lane). Under the guidance of protein maker, corresponding bands of CD63, TSG101 and calnexin is cut and transferred to a PVDF membrane. After blocking with BSA, the membrane is incubated with antibodies, including rabbit polyclonal antibody CD63 (sc-5275, Santa Cruz, CA, USA), TSG101 (sc-13,611, Santa Cruz, CA, USA), Alix (sc-53,540, Santa Cruz, CA, USA) and calnexin (10,427–2-AP, Promega, Madison, WI). Finally, the membrane is rinsed to eliminate unbounded antibodies and visualized on the Tanon4600 chemiluminescence image analysis system (Tanon, Shanghai, China).
RNA extraction and qPCR
The predictors used in the development of this multivariable model in this study were the expression level of RNA candidates (i.e., TK1, CDK1, MYBL2, TPX2, FOXM1, UBE2C, BIRC5, CDC20, MMP11, and CCNB1.). In order to quantitatively analyze the expression of these candidate markers, EVs RNA is extracted by miRNeasy Mini Kit (217004, Qiagen, German) following standard protocol. The resultant RNA product is subjected to reverse transcription and qPCR using PrimeScript RT reagent Kit (RR037, Takara, Japan) and Premix Ex Taq master mix (RR390A, Takara, Japan) respectively. All primers and probes (Table S1) are synthesized by Synbio Technologies (Suzhou, China). External reference made from cell line RNA is added in each plate for data rectification. Finally, ΔCT for each individual marker is obtained after normalization by housekeeping gene.
Statistical analysis
All statistical analysis is performed using R language (R 3.5.1, www.r-project.org). Based on The Cancer Genome Atlas (TCGA) database, differentially expressed genes (DEGs) of bladder cancer and para-carcinoma were identified. The significance of the difference between the two groups was calculated by Mann Whitney u-test. The fold change (FC) between the two groups was calculated according to the median expression. The threshold of DEGs was set as significance P value <0.01, and FC >2. The expression correlations for selected genes were calculated to remove redundant genes with high correlation.
To build the diagnostic model, ΔCT value for each marker is obtained through normalization by internal housekeeping gene. Logistic regression model was established based on the values of ΔCT. In the case that the CT value is not available in a PCR assay because the concentration of target RNA molecules was too low to detect, the value of ΔCT would be set to 20 to facilitate subsequent analysis process.
Result
Schematic illustration of the workflow
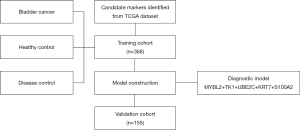
The schematic illustration of the workflow is shown in Figure 1. First, RNA-seq data of 408 cancer and 19 para-carcinoma tissues of bladder cancer patients are downloaded from The Cancer Genome Atlas (TCGA) dataset. Through differential gene expression analysis, a panel of 12 candidate genes was identified. The expression level and diagnostic performance of these candidate genes are then evaluated by a training cohort of 368 clinical samples. The optimal diagnostic model including five mRNA markers (i.e., MYBL2, TK1, UBE2C, KRT7, and S100A2) is further verified by a validation cohort of 155 samples.
Comparison of mRNA and miRNA as candidate markers
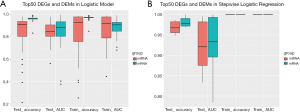
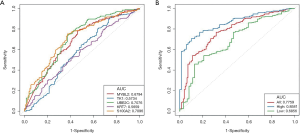
The detection of bladder cancer by mRNA and miRNA had been both reported before. To determinate which one of them is likely to yield a more promising performance, we first carried out a comparison in tissue level with TCGA dataset (Table 1). Briefly, a total of 7,053 differentially expressed gene (DEGs) and 297 differential expressed miRNA (DEMs) between bladder cancer and para-carcinoma tissue were identified and listed in descending order by their significant level. The expression level of a given gene is determined by its FPKM (Fragments per Kilobase Million) value. Top 50 mRNAs and miRNAs were selected from each list. The performance of either a single marker or marker combination was evaluated basing on logistic regression model. For a single marker, the data set was randomly divided into a training set and a testing set before analysis. For a marker combination, DEGs and DEMs were analyzed respectively with stepwise logistic regression model. The training set and testing set was randomly divided and repeated for ten times. The accuracy and AUC of a single marker (Figure 2A) or marker combination (Figure 2B) were evaluated. Apparently, DEGs was less sensitive to random grouping comparing to DEMs, thus providing a more stable result. Therefore, we decided to focus on DEGs in the following study.
Full table
Screening of candidate markers
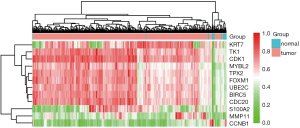
In this step, two criteria were applied to identify candidate markers from existed DEGs: (I) FC of gene expression between bladder cancer and para-carcinoma >8, (II) FC of gene expression between bladder cancer and prostate/renal cancer >5. As a result, a total of 10 markers were selected, which were TK1, CDK1, MYBL2, TPX2, FOXM1, UBE2C, BIRC5, CDC20, MMP11, and CCNB1. Furthermore, to ensure the diagnostic performance in low grade bladder cancer, two additional markers KRT7 and S100A2, both of which were significantly elevated in low grade bladder cancer comparing to normal and para-carcinoma tissues, were incorporated in the following study. The expression levels of all 12 candidate markers in 427 samples were showed in Figure 3. Owing to relatively stable expression, SLC25A6 was chosen as internal reference.
Isolation and characterization of EVs
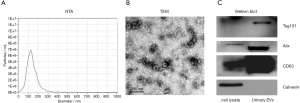
The size and morphology of urinary EVs isolated by PEG precipitation was characterized by nanoparticle tracking analysis (NTA) and transmission electron microscope (TEM). NTA result suggested the size distribution of urinary EVs ranged from 50 to 200 nm, with the peak value in 124.2 nm (Figure 4A). As shown in Figure 4B, the obtained EVs were either oval or bowl-shape structured without nucleus under TEM. EVs associated protein markers, CD63, Alix and TSG101 can all be detected in western blot analysis, while the negative marker calnexin was absent (Figure 4C). The above results were consistent with published literature, indicating good quality of isolated EVs (16).
Model establishment
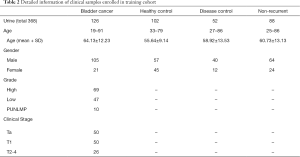
Urinary EVs isolated from 126 bladder cancer patients, 102 healthy controls, 52 disease controls, (i.e., 9 patients with benign prostatic hyperplasia, 25 with cystitis, 8 with non-bladder urothelial carcinoma, 10 with urinary calculus), and 88 non-recurrent bladder cancer patients were used for analysis. Detailed information of clinical samples enrolled in training cohort, such as age, gender, grade, and clinical stage were listed in Table 2. It is worthy to mention that the abundance of mRNA MMP11 and CCNB1 were lower than the minimum detection level in over a half of the samples, thus were excluded from the subsequent analysis. The ΔCT of the rest RNA markers were calculated by subtracting the CT value of housekeeping gene SLC25A6. Taking cystoscopy as the gold standard, the diagnostic model of each marker was established and analyzed. Eventually, five mRNAs MYBL2, TK1, UBE2C, KRT7, and S100A2 with relatively better performance and more evident differential expression were chosen as the final marker panel (ExoPanel).
Full table
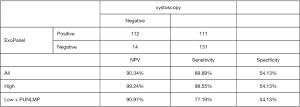
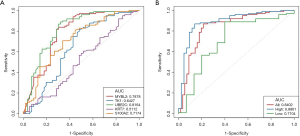
The receiver operating characteristic (ROC) curve of the five markers MYBL2, TK1, UBE2C, KRT7 and S100A2 were showed in Figure 5A. The overall area under the curve (AUC) of top 2 markers, S100A2 and UBE2C, were both very close to 0.71. For ExoPanel, the AUC was 0.7759 (95% CI: 0.7259−0.8260) in all samples, 0.8581 (95% CI: 0.8147−0.9014) in the high-level samples and 0.6856 (95% CI: 0.6065−0.7647) in the low-level samples (Figure 5B). As showed in Figure 6, a 2×2 table analysis was carried out to compare the performance of ExoPanel and cystoscopy in this training corhort. As a supplementary measure, one potential application of this assay is to rule out low risk patient and ensure those in real need receive cystoscopy examination timely. To serve this purpose, the priority was given to negative predictive value (NPV) over other index during model construction. In this case, the predictive value for each participant was calculated as Probability = EXP(Z)/(1+EXP(Z)), while Z = a+b *ΔCT MYBL2 + c*ΔCT TK1 + d*ΔCTUBE2C + e*ΔCTKRT7 +f*ΔCTS100A2 (a= 0.231428866732931, b=−0.0616939015948833, c=0.484404962244116, d=−0.307187880933284, e=−0.0268137601118776, f=–0.225814663743345). The cutoff value was determinated as 0.241942833741954. If the predictive value for a participant is higher than cutoff, he or she would be assessed as positive by ExoPanel, or otherwise negative. As a result, the overall NPV, SN and SP of the final model were 90.34% (95% CI: 84.04–94.42%), 88.89% (95% CI: 81.75–93.57%) and 54.13% (95% CI: 47.63–60.50%). In particular, the SN of the model was 98.55% (95% CI: 95.65–100%) in high grade bladder cancers and 77.19% (95% CI: 66.67–87.72%) in low grade bladder cancers. Among 128 bladder cancer samples confirmed by cystoscopy, about 14 cases (1 high-grade and 13 low-grade bladder cancer) were diagnosed as negative by ExoPanel.
Model validation
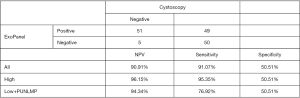
To validate the performance of established model, 56 bladder cancer patients, 50 healthy controls, 12 disease controls, (i.e., 4 patients with benign prostatic hyperplasia, 5 with cystitis, 2 with non-bladder urothelial carcinoma, 2 with urinary calculus), and 37 non-recurrent bladder cancer patients were enrolled as training cohort. Detailed information, such as age, gender, grade, and clinical stage were listed in Table 3. EVs isolated from their urine were subjected to qPCR analysis following the same protocol as above. In good accordance with training cohort, the AUC in validation cohort was round 0.8402 (95% CI: 0.7690–0.9114) for all samples, 0.8901 (95% CI: 0.8262–0.9539) for high-grade bladder cancers, and 0.7104 (95% CI: 0.5331–0.8877) for low-grade bladder cancers (Figure 7). A 2×2 table analysis was used to compare the performance of ExoPanel and cystoscopy in this validation cohort (Figure 8). Except for two high-grade and 3 low-grade bladder cancers, 51 out of 56 patients were consistent with cystoscopy. The overall NPV, SN and SP were 90.91% (95% CI: 79.29–96.60%), 91.07% (95% CI: 79.63–96.67%) and 50.51% (95% CI: 40.34–60.63%). For low-grade bladder cancers in particular, NPV, SN and SP of ExoPanel were 94.34% (95% CI: 83.37–98.53%), 76.92% (95% CI: 53.85–100%) and 50.51% (95% CI: 40.40–60.61%). This result suggested that the established model can be used to predict patients’ disease status.
Full table
Discussion
As supplementary means to cystoscopy, a number of urinary based tests were developed for non-invasive diagnosis of bladder cancer. For example, urine cytology, FISH (Vysis), NMP22, and BTA test had been put to use for routine clinical practice (17,18). Urine cytology and FISH (Vysis), two assays based on exfoliated cells, still had difficulty in diagnosing low-grade bladder cancers. By detecting NMP22 released by apoptotic cells, this assay had a SP 70–83% and SN 62–75%. BTA test, which detect bladder cancer by qualitatively analyzing the urinary human complement factor H-related protein (hcfHrp), showed a SP 64–82% and SN 54–69%. However, both of them would be affected by benign diseases, such as hematuria, infection, hyperplasia, or cystitis, and lead to false positive. This might be attributed to the intrinsic disadvantage of using a single marker. Theoretically, it is possible to improve the anti-interference ability by incorporating the synergistic effect of additional markers.
In recent years, our knowledge of EVs has developed rapidly (19,20). EVs can be found in so many types of bio-fluids, and they also carry a variety of parental cell derived bio-molecules. Therefore, it is an ideal bio-marker library and emerged as an important analyte in liquid biopsy. Given that bladder cancer cell derived EVs can be directly released and accumulate in the urine, it is possible to detect bladder cancer by analyzing urinary EVs. To avoid the interference by other diseases, the selected EVs markers must be highly specific for bladder cancers. Due to this reason, we not only consider the differential expression level between cancer and para-carcinoma tissue, but also take other urinary system related diseases into account.
As mentioned previously, instead of performing RNA-seq with urinary EVs, we chose an alternative way that is to identify potential RNA markers by analyzing tissue data downloaded from TCGA. Of course, one thing must be taken into consideration is that the absolute abundance of these markers in urinary EVs is different from tissue. In our case, for example, the copy numbers of two candidate markers MMP11 and CCNB1 in urinary EVs were too low to be detected by qPCR in over a half of samples. It suggests that the expression level is a crucial factor in liquid biopsy, and extra attention should be paid during marker screening process. Interestingly, unlike cytology analysis, which prefers second morning urine because exfoliated cells might be damaged after prolonged immersion in the urine, collecting the first morning urine for EVs based assay can slightly increase the total RNA amount.
As expected, the diagnostic performance of 5 mRNA panel is remarkably better than any one of them, indicating that there might be synergistic effect between different genes. Evidence in favor of this hypothesis can be found in the biological functions of these genes. For example, although three out of five markers (MYBL2, TK1 and UBE2C) were associated with cell cycle progression, up regulation of these mRNA are likely to correspond to different cell cycle (21-23). Known as a member of keratin gene family, altered expression of KRT7 might be related to the neoplastic process of urothelium in early phase of bladder cancer (24). S100A2 played an important role in cytoskeleton organization, as well as differentiation and regeneration of tissue. Its expression was found to be reduced in several types of cancer and hypothesized as a potential tumor suppressor. In bladder cancer, however, overexpression of S100A2 was also regarded as an early tumorigenic event (25).
Considering that the main purpose of this assay is to accurately screen out patients that in real need of cystoscopy inspection, the priority was first given to NPV, then SN. Thus, SP was compromised to ensure the high value of NPV and SN. As a result, the overall performance of ExoPanel was NPV=90.91% (95% CI: 79.29–96.60%), SN=91.07% (95% CI: 79.63–96.67%), and SP=50.51% (95% CI: 40.34–60.63%). In high-grade samples, the number were NPV=96.15% (95% CI: 85.67–99.33%), SN=95.35% (95% CI: 88.37–100%). And in low-grade sample, the performance were NPV= 94.34% (95% CI: 83.37–98.53%), and SN=76.92% (95% CI: 53.85–100%). However, this study has several limitations. First, this study is not double blind, and all samples were collected from one center. Besides, the sample size, especially validation cohort is still small. Finally, no follow up study was performed for those patients with inconsistent results between cystoscopy and ExoPanel.
Conclusions
In this paper, we had introduced a method to identify potential EVs RNA marker from the RNA-seq data of tissues. Following this method, a five mRNA model ExoPanel was established, which showed promising performances in the diagnosis of bladder cancer. This assay can identify a significant amount of negative patients before carrying out cystoscopy test. By doing so, it is possible to reduce the unnecessary operation of cystoscopy, improve patients’ quality of life, and reduce the lifetime treatment cost per person.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1057
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1057
Peer Review File: Available at http://dx.doi.org/10.21037/tau-20-1057
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1057). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (revised in 2013). This study was approved by the ethics committee of Chinese PLA General Hospital (C2017-037-01). Informed consents are obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Richters A, Aben KKH, Kiemeney L. The global burden of urinary bladder cancer: an update. World J Urol 2020;38:1895-904. [Crossref] [PubMed]
- Jocham D, Stepp H, Waidelich R. Photodynamic diagnosis in urology: state-of-the-art. Eur Urol 2008;53:1138-48. [Crossref] [PubMed]
- Burke DM, Shackley DC, O'Reilly PH. The community-based morbidity of flexible cystoscopy. BJU Int 2002;89:347-9. [Crossref] [PubMed]
- Johnson MI, Merrilees D, Robson WA, et al. Oral ciprofloxacin or trimethoprim reduces bacteriuria after flexible cystoscopy. BJU Int 2007;100:826-9. [Crossref] [PubMed]
- Renshaw AA, Granter SR. Fine needle aspiration of chromophobe renal cell carcinoma. Acta Cytol 1996;40:867-72. [Crossref] [PubMed]
- Gregoire M, Fradet Y, Meyer F, et al. Diagnostic accuracy of urinary cytology, and deoxyribonucleic acid flow cytometry and cytology on bladder washings during followup for bladder tumors. J Urol 1997;157:1660-4. [Crossref] [PubMed]
- Dimashkieh H, Wolff DJ, Smith TM, et al. Evaluation of urovysion and cytology for bladder cancer detection: a study of 1835 paired urine samples with clinical and histologic correlation. Cancer Cytopathol 2013;121:591-7. [Crossref] [PubMed]
- Mohrmann L, Huang HJ, Hong DS, et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin Cancer Res 2018;24:181-8. [Crossref] [PubMed]
- Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177-82. [Crossref] [PubMed]
- Min L, Zhu S, Chen L, et al. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J Extracell Vesicles 2019;8:1643670. [Crossref] [PubMed]
- Lin SY, Chang CH, Wu HC, et al. Proteome Profiling of Urinary Exosomes Identifies Alpha 1-Antitrypsin and H2B1K as Diagnostic and Prognostic Biomarkers for Urothelial Carcinoma. Sci Rep 2016;6:34446. [Crossref] [PubMed]
- Juracek J, Peltanova B, Dolezel J, et al. Genome-wide identification of urinary cell-free microRNAs for non-invasive detection of bladder cancer. J Cell Mol Med 2018;22:2033-8. [Crossref] [PubMed]
- Berrondo C, Flax J, Kucherov V, et al. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS One 2016;11:e0147236. [Crossref] [PubMed]
- Murakami T, Yamamoto CM, Akino T, et al. Bladder cancer detection by urinary extracellular vesicle mRNA analysis. Oncotarget 2018;9:32810-21. [Crossref] [PubMed]
- Rider MA, Hurwitz SN, Meckes DG Jr. ExtraPEG: A Polyethylene Glycol-Based Method for Enrichment of Extracellular Vesicles. Sci Rep 2016;6:23978. [Crossref] [PubMed]
- Maas M, Bedke J, Stenzl A, et al. Can urinary biomarkers replace cystoscopy? World J Urol 2019;37:1741-9. [Crossref] [PubMed]
- Simon MA, Lokeshwar VB, Soloway MS. Current bladder cancer tests: unnecessary or beneficial? Crit Rev Oncol Hematol 2003;47:91-107. [Crossref] [PubMed]
- De Toro J, Herschlik L, Waldner C, et al. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front Immunol 2015;6:203. [Crossref] [PubMed]
- Kalluri R. The biology and function of exosomes in cancer. J Clin Invest 2016;126:1208-15. [Crossref] [PubMed]
- Dastsooz H, Cereda M, Donna D, et al. A Comprehensive Bioinformatics Analysis of UBE2C in Cancers. Int J Mol Sci 2019;20:2228. [Crossref] [PubMed]
- Zhou J, He E, Skog S. The proliferation marker thymidine kinase 1 in clinical use. Mol Clin Oncol 2013;1:18-28. [Crossref] [PubMed]
- Musa J, Aynaud MM, Mirabeau O, et al. MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis 2017;8:e2895. [Crossref] [PubMed]
- Aaboe M, Marcussen N, Jensen KM, et al. Gene expression profiling of noninvasive primary urothelial tumours using microarrays. Br J Cancer 2005;93:1182-90. [Crossref] [PubMed]
- Yao R, Lopez-Beltran A, Maclennan GT, et al. Expression of S100 protein family members in the pathogenesis of bladder tumors. Anticancer Res 2007;27:3051-8. [PubMed]