Identification of a potentially functional circRNA-miRNA-mRNA ceRNA regulatory network in bladder cancer by analysis of microarray data
Introduction
Bladder cancer (BCa) is the most common malignant tumor of the genitourinary system, with high mortality rates worldwide (1). BCa is categorized into two subtypes, including non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC). Most BCa patients are primarily diagnosed with NMIBC. However, approximately 50–70% of patients with NMIBC will experience disease recurrence within 5 years. Also, 10–30% of patients will develop MIBC, which currently has a poor prognosis (2). Although several therapeutic approaches, including surgical resection, chemotherapy, and radiotherapy, have been developed, the 5-year survival rate of MIBC patients is only approximately 60% (3,4). It is of vital importance to identify novel biomarkers and effective targets for BCa.
Circular RNAs (circRNAs) are a novel class of RNAs that are generated via endogenous splicing. These covalently closed-loop structures are resistant to exonuclease cleavage due to the absence of 5'-3' polyadenylation tails. The high stability displayed by circRNAs suggests they are strong candidates, as biomarkers (5,6). There is evidence that circRNAs play critical roles in cancer biology (7-9), and more specifically, in the development of BCa. One mechanism employed by circRNAs is via competition with microRNAs (miRNA). The competing endogenous circRNAs, known as ceRNAs, serve as miRNA sponges. For instance, a highly conserved circRNA, ciRS-7 (also termed CDR1as), exerts anti-tumor functions in BCa by sponging miR-135a (10). CircHIPK3 can effectively inhibit tumor aggression and metastasis of BCa by acting as ceRNAs of miR-558 (11). CircRIP2 accelerates BCa progression by targeting the miR-1305/Tgf-β2/smad3 axis (12). Meanwhile, CircFAM114A2 suppresses BCa progression via regulating ∆NP63 by sponging miR-762 (13). Circular RNA circPICALM inhibits BCa metastasis via influencing FAK phosphorylation by acting as ceRNAs of miR-1265 (14). These findings all together strongly suggest the importance of circRNAs as novel biomarkers in the regulation and development of BCa. However, many unidentified circRNAs remain to be investigated.
In this study, a circRNA microarray from the GEO database was used to screen differentially expressed circRNAs between BCa tissues and paired-matched adjacent normal tissues. To depict whether the differentially expressed circRNAs functioned as ceRNAs in BCa, we predicted their sponged miRNAs and miRNAs-targeted mRNAs. The signaling pathways of miRNA-targeted mRNAs were then analyzed, and a circRNA-miRNA-mRNA regulatory network was constructed. A protein-protein interaction (PPI) network was subsequently established, and the hub genes were subsequently identified. Our results provide significant insights into the molecular mechanisms that regulate the progression of BCa from the circRNA-miRNA-mRNA network. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-660).
Methods
Data collection
The gene expression profiles of BCa were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/) database. The miRNA and mRNA data of BCa were downloaded from TCGA (https://portal.gdc.cancer.gov). The circRNA expression profile of GSE92675 was obtained from the GEO database, including 4 pairs of matched BCa (GSM2434319, GSM2434321, GSM2434323, GSM2434325) and para-cancer tissues (GSM2434320, GSM2434322, GSM2434324, GSM2434326). A total of 433 mRNA samples (414 BCa samples and 19 normal bladder samples) were obtained from the TCGA database.
Differentially expressed circRNAs and mRNAs
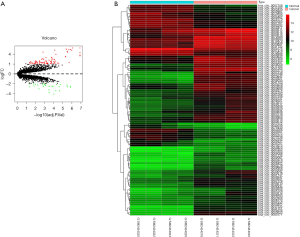
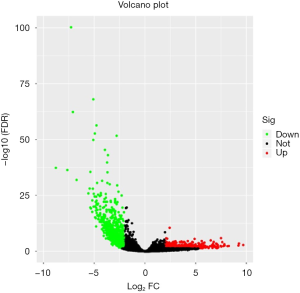
The Limma package was used to identify the differentially expressed circRNAs between BCa tissue and the adjacent normal tissue from the GEO database (FDR value <0.05 and |log2FC| >2). Additionally, we used the EdgeR package to screen the differentially expressed mRNAs with thresholds of |log2FC| >2 and P value <0.05.
Prediction of circRNA-miRNA binding sites
Prediction of interactions between circRNAs and miRNAs was conducted using the CircInteractome database. MiRNAs with a context score percentile >98 were eventually selected.
Screening of miRNA-targeted mRNAs and pathway analysis
The miRNA-targeted mRNAs were predicted with the TargetScan prediction tool and miRDB software. The signaling pathways of miRNA-targeted mRNAs were analyzed by Enrichr software. The effect of miRNAs on the survival of BCa patients was analyzed by using the online Kaplan-Meier Plotter database.
Construction of a circRNA-miRNA-mRNA regulatory network
The ceRNA regulatory network was established on the foundation of the interactions between the differentially expressed circRNAs, the predicted miRNAs, and the differentially expressed mRNAs. Cytoscape 3.7.1 software was used to visualize the established ceRNA regulatory network.
Establishment of PPI regulatory network and identification of hub genes
A PPI network was constructed by using the STRING (V 11.5) and visualized by the CytoCope 3.7.1 software. Additionally, the highly interacted hub genes from the PPI network were screened by the CytoHubba plugin in Cytoscape.
Verification of hub gene expression with the GEPIA database
To further analyze the expression level of hub genes mRNA in BCa. We explored the mRNA expression level of these hub genes in the web-based GEPIA database (http://gepia.cancer-pku.cn/) with the settings P≤0.05 and |log2(FC)| ≥1.
Statistical analysis
All statistical analyses were performed using R (v.3.6.0) software and SPSS v.24.0 software (IBM Corp.). The correlation between miRNA expression and patients’ survival was assessed by Kaplan-Meier method. The expression of hub genes in different tissues was analyzed by Student’s t-test. The P value <0.05 was considered statistically significant.
Results
Identification of differentially expressed circRNAs
The GSE92675 dataset from GEO was analyzed by the limma package in R (P value <0.05 and log2|fold change| >2), we identified 79 up-regulated and 28 down-regulated circRNAs (Figure 1). Also, a total of 1,229 differential expressed mRNAs (472 up-regulated and 757 down-regulated mRNAs) were identified from the TCGA database between BCa tissues and normal bladder tissues (FDR value <0.05 and log2|fold change| >2) (Figure 2). Subsequently, three circRNAs were selected according to the P value. The basic structure patterns of these three circRNAs (has_circ_0023642, has_circ_0047322, has_circ_0041151) are exhibited in Figure 3.

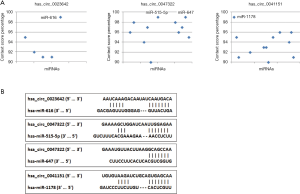
Identification of three circRNA-miRNA interactions
Accumulated evidence has demonstrated that some circRNAs derived from exons play crucial roles in tumor progression by acting as miRNAs “decoys” to sponge miRNAs. To identify whether these three circRNAs performed similar roles in BCa, we carried out a context score percentile screening in the CircInteractome database. Four miRNAs (miR-616, miR-515-5p, miR-647, miR-1178) with a context score percentile >98 were selected (Figure 4A). The specific binding sites between circRNAs and miRNAs were shown in Figure 4B. Enrichr was employed to explore the signaling pathways of these four miRNAs. As shown in Figure 5, all these four miRNAs are strongly associated with some cancer-related pathways.
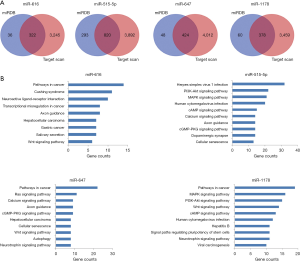
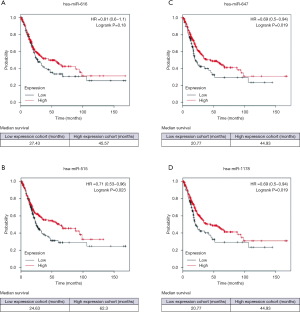
Expression of the four miRNAs with the Kaplan-Meier Plotter database
The significance of the selected miRNA expression in BCa was further explored. We analyzed the effect of miRNA expression on the survival of BCa patients by using the online Kaplan-Meier Plotter database. As shown in Figure 6, the overall survival of BCa patients who expressed high-levels of miR-515-5p, miR-647, and miR-1178 exhibited better survival compared to patients with low miRNA expression. The median survival of BCa patients with high expression of miR-616 (45.57 months) was higher than the patients with low miR-616 expression (27.43 months). However, this difference is not statistically significant (P=0.18). These results suggest that these miRNAs might play potential tumor-suppressive functions in BCa.
Construction of circRNA-miRNA-mRNA regulatory networks in BCa
Ten thousand and seventy-one target genes of the four miRNAs mentioned above were obtained from the TargetScan prediction tool. Additionally, 472 up-regulated genes in BCa were identified from TCGA (Figure 2). These investigations led to the identification of 181 overlapping target genes that played critical roles in BCa (Figure 7A).
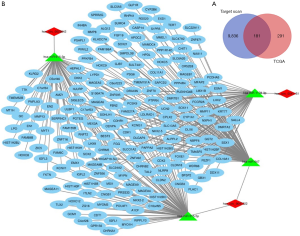
A circRNA-miRNA-mRNA network was constructed through integrating the circRNA-miRNA interactions and miRNA-mRNA interactions (Figure 7B). The construction of the network provided preliminary insight into the links between the differential circRNAs (has_circ_0023642, has_circ_0047322, has_circ_0041151), the four miRNAs (miR-616, miR-515-5p, miR-647, miR-1178) and the 181 mRNAs.
Construction of PPI network
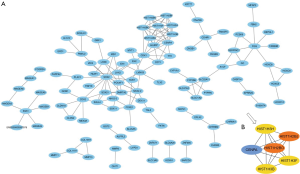
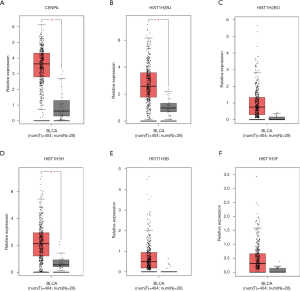
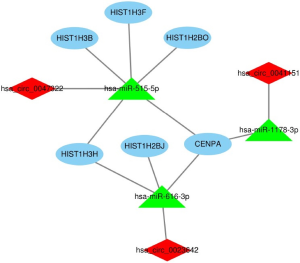
After removing unconnected nodes, a PPI network consisting of 106 nodes and 160 edges were constructed to view the interactions among the 181 mRNAs (Figure 8A). To explore the key circRNA-miRNA-mRNA regulatory modules in the progression of BCa, we used the MCODE software to screen hub genes from the PPI network. The significant module containing 6 nodes and 15 edges was identified (Figure 8B). These hub genes were CENPA, HIST1H2BJ, HIST1H2BO, HIST1H3H, HIST1H3B, HIST1H3F (Figure 8B). The expression levels of these six genes in BCa were shown in Figure 9. A circRNA-miRNA-hub genes sub-network was then built to delineate the links among the differential circRNAs, miRNA and hub genes (Figure 10), including has_circ_0047322-hsa-miR-515-5p-HIST1H3B regulatory axis, has_circ_0047322-hsa-miR-515-5p-HIST1H3F regulatory axis, has_circ_0047322-hsa-miR-515-5p-HIST1H2BO regulatory axis, has_circ_0047322-hsa-miR-515-5p-HIST1H3H regulatory axis, has_circ_0047322-hsa-miR-515-5p-CENPA regulatory axis, has_circ_0023642-hsa-miR-616-3p-HIST1H3H regulatory axis, has_circ_0023642-hsa-miR-616-3p-HIST1H2BJ regulatory axis, has_circ_0023642-hsa-miR-616-3p-CENPA regulatory axis, has_circ_0041151-hsa-miR-1178-3p-CENPA regulatory axis, were found from the network.
Discussion
Accumulating evidence suggests that circRNAs are abundant and stable in eukaryotic cells, demonstrating high conservation and disease specificity (15-17). It has been reported that circRNAs that are abnormally expressed play crucial roles in various cancers (18-22). Their resistance to exonuclease cleavage due to their covalently closed structure has resulted in their routine use as prognostic and diagnostic biomarkers. In BCa, various circRNAs, including Cdr1as (10), circHIPK3 (11), circRIP2 (12), circFAM114A2 (13), circPICALM (14), circZKSCAN1 (23), circACVR2A (24), circDOCK1 (25), have been reported to exert crucial roles in regulating the disease progression. CircRNAs have also been used to aid in the clinical diagnosis of BCa and subsequent courses of therapy. However, many circRNAs remain unidentified.
In this study, microarray data were collected from the GEO database. Three up-regulated circRNAs (has_circ_0023642, has_circ_0047322, has_circ_0041151) were subsequently screened in BCa for further analysis. To ascertain whether those above three circRNAs function as miRNA sponges in BCa, the CircInteractome online tool was utilized to predict miRNAs with binding potential to circRNAs. Some circRNAs harbor abundant miRNA binding sites, thereby acting as “decoys” or “sponges” to regulate gene expression. Four miRNAs (miR-616, miR-515-5p, miR-647, miR-1178) were determined to have high binding potentials with these three circRNAs. We then analyzed the effect of the miRNAs on the survival of BCa patients via the online Kaplan-Meier Plotter database. According to the results, three miRNAs, including miR-515-5p, miR-647, and miR-1178, showed a positive association with the better survival of BCa, which suggested their potential tumor-suppressive functions in BCa.
Following the collection of the 181 overlapping genes between the up-regulated genes from TCGA and the target genes of the four miRNAs in BCa, we constructed a circRNA-miRNA-mRNA regulatory network. We found that has_circ_0023642, has_circ_0047322, has_circ_0041151 may act as miRNA sponges to capture miR-616, miR-515-5p, miR-647, miR-1178, and subsequently regulate the mRNA expression of the 181 overlapping genes. Our results provide clinical evidence of the ceRNA regulatory mechanism of has_circ_0023642, has_circ_0047322, and has_circ_0041151 in BCa. To further demonstrate the action of the ceRNA network, we subsequently constructed a PPI network, screening six hub genes, including CENPA, HIST1H2BJ, HIST1H2BO, HIST1H3H, HIST1H3B, HIST1H3F. The important roles of these six genes in cancer have also been demonstrated previously (26-30). In our study, we identified nine circRNA-miRNA-mRNA axes (has_circ_0047322-hsa-miR-515-5p-HIST1H3B regulatory axis, has_circ_0047322-hsa-miR-515-5p-HIST1H3F regulatory axis, has_circ_0047322-hsa-miR-515-5p-HIST1H2BO regulatory axis, has_circ_0047322-hsa-miR-515-5p-HIST1H3H regulatory axis, has_circ_0047322-hsa-miR-515-5p-CENPA regulatory axis, has_circ_0023642-hsa-miR-616-3p-HIST1H3H regulatory axis, has_circ_0023642-hsa-miR-616-3p-HIST1H2BJ regulatory axis, has_circ_0023642-hsa-miR-616-3p-CENPA regulatory axis, has_circ_0041151-hsa-miR-1178-3p-CENPA regulatory axis), indicating competitive regulatory relationships of has_circ_0023642, has_circ_0047322 and has_circ_0041151 with the six hub genes in BCa. However, further in-depth studies are needed to verify the possible roles of these nine axes in BCa.
Acknowledgments
Funding: Natural Science Foundation of Hebei Province of China (H2020206109).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-660
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-660). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Antoni S, Ferlay J, Soerjomataram I, et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol 2017;71:96-108. [Crossref] [PubMed]
- Fujii Y. Prediction models for progression of non-muscle-invasive bladder cancer: A review. Int J Urol 2018;25:212-8. [Crossref] [PubMed]
- Luke C, Tracey E, Stapleton A, et al. Exploring contrary trends in bladder cancer incidence, mortality and survival: implications for research and cancer control. Intern Med J 2010;40:357-62. [Crossref] [PubMed]
- Racioppi M, D’Agostino D, Totaro A, et al. Value of current chemotherapy and surgery in advanced and metastatic bladder cancer. Urol Int 2012;88:249-58. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014;20:1829-42. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett 2017;396:138-44. [Crossref] [PubMed]
- Li P, Yang X, Yuan W, et al. CircRNA-Cdr1as Exerts Anti-Oncogenic Functions in Bladder Cancer by Sponging MicroRNA-135a. Cell Physiol Biochem 2018;46:1606-16. [Crossref] [PubMed]
- Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep 2017;18:1646-59. [Crossref] [PubMed]
- Su Y, Feng W, Shi J, et al. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-β2/smad3 pathway. Mol Cancer 2020;19:23. [Crossref] [PubMed]
- Liu T, Lu Q, Liu J, et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating ∆NP63 by sponging miR-762. Cell Death Dis 2020;11:47. [Crossref] [PubMed]
- Yan D, Dong W, He Q, et al. Circular RNA circPICALM sponges miR-1265 to inhibit bladder cancer metastasis and influence FAK phosphorylation. EBioMedicine 2019;48:316-31. [Crossref] [PubMed]
- Santer L, Bär C, Thum T. Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Mol Ther 2019;27:1350-63. [Crossref] [PubMed]
- Garikipati VNS, Verma SK, Cheng Z, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun 2019;10:4317. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Sang Y, Chen B, Song X, et al. circRNA_0025202 Regulates Tamoxifen Sensitivity and Tumor Progression via Regulating the miR-182-5p/FOXO3a Axis in Breast Cancer. Mol Ther 2019;27:1638-52. [Crossref] [PubMed]
- Hong X, Liu N, Liang Y, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer 2020;19:33. [Crossref] [PubMed]
- Wu G, Zhou W, Pan X, et al. Circular RNA Profiling Reveals Exosomal circ_0006156 as a Novel Biomarker in Papillary Thyroid Cancer. Mol Ther Nucleic Acids 2020;19:1134-44. [Crossref] [PubMed]
- Chen W, Quan Y, Fan S, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett 2020;475:119-28. [Crossref] [PubMed]
- Sun X, Wang M, Xu R, et al. Prognostic model based on circular RNA circPDK1 for resected lung squamous cell carcinoma. Transl Lung Cancer Res 2019;8:907-19. [Crossref] [PubMed]
- Bi J, Liu H, Dong W, et al. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer 2019;18:133. [Crossref] [PubMed]
- Dong W, Bi J, Liu H, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer 2019;18:95. [Crossref] [PubMed]
- Liu P, Li X, Guo X, et al. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif 2019;52:e12614. [Crossref] [PubMed]
- Liu WT, Wang Y, Zhang J, et al. A novel strategy of integrated microarray analysis identifies CENPA, CDK1 and CDC20 as a cluster of diagnostic biomarkers in lung adenocarcinoma. Cancer Lett 2018;425:43-53. [Crossref] [PubMed]
- Takada M, Zhang W, Suzuki A, et al. FBW7 Loss Promotes Chromosomal Instability and Tumorigenesis via Cyclin E1/CDK2-Mediated Phosphorylation of CENP-A. Cancer Res 2017;77:4881-93. [Crossref] [PubMed]
- Li X, Tian R, Gao H, et al. Identification of a histone family gene signature for predicting the prognosis of cervical cancer patients. Sci Rep 2017;7:16495. [Crossref] [PubMed]
- Xie W, Zhang J, Zhong P, et al. Expression and potential prognostic value of histone family gene signature in breast cancer. Exp Ther Med 2019;18:4893-903. [Crossref] [PubMed]
- Mirisola V, Mora R, Esposito AI, et al. A prognostic multigene classifier for squamous cell carcinomas of the larynx. Cancer Lett 2011;307:37-46. [Crossref] [PubMed]
(English Language Editors: E. Tan and J. Chapnick)