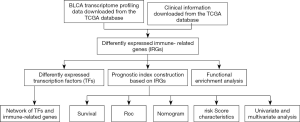
Prognostic model of 10 immune-related genes and identification of small molecule drugs in bladder urothelial carcinoma (BLCA)
Introduction
Bladder cancer (BC), as one of the major causes of cancer related mortality worldwide, it attributed to nearly 549,000 new cases and 200,000 deaths in the United States, according to Global Cancer Statistics 2018 (1). Tobacco smoking and occupational exposure to certain chemical carcinogens remained the main risk factors of BC (2-4). Because of the gradual popularization of bladder B-ultrasound, transurethral cystoscopy, etc., many BC patients could be diagnosed in early time and receive timely and effective treatment, including transurethral resection, radiation therapy, and chemotherapy. However, the prognosis for BC patients was still unsatisfactory, due to frequent recurrence and metastasis (5,6).
A growing number of evidence had highlighted the important roles of immune-related genes (IRGs) in cancer initiation, progression, metastasis and so on. Therein, CD58, a cell-surface glycosylated adhesion molecule, could enhance the self-renewal of colorectal tumor-initiating cells and promote epithelial-mesenchymal transition (EMT) process through activating Wnt/β-catenin pathway in colorectal cancers (7). The overexpression of ZBTB20, a transcriptional repressor, could lead to enhancement of migration and invasion ability of gastric cancer by inhibiting IκBα to activate NF-κB (8). Besides, ZFP36L2, a RNA-binding protein, had been shown to inhibit cell proliferation in pancreatic ductal adenocarcinoma (PDAC), and it was associated with good prognosis of PDAC patients (9). IRGs also played very important roles in BC. Martínez et al. demonstrated that BMP-4, secreted by BC cells, induced monocyte/macrophage polarization toward an M2 phenotype, and promoted tumor progression (10). Ramakrishnan et al. found that inhibition of EZH2 in muscle-invasive BC with KDM6A and SWI/SNF family member mutations could activate a natural killer (NK) cell-based immune response to drive tumor differentiation and death in BC cells and xenografts (11). Cheah et al. elucidated the importance of CD14 in BC, and showed that high levels of CD14 resulted in the increase of inflammation mediators and drove tumor proliferation to promote tumor growth (12). Although a number of previous studies had proposed the roles of IRGs in BC, there were few focusing on their importance to systematically predict overall survival (OS) of BC patients.
As a result, we aimed to establish a signature based on IRGs that could predict OS for BC patients in this study. We utilized the transcriptome data downloaded from The Cancer Genome Atlas (TCGA) to develop a prognostic index (PI) by using univariate Cox regression analysis and least absolute shrinkage and selection operator (LASSO) regression analysis. To evaluate the clinical values of this PI, we analyzed whether this signature was associated with the survival outcome of BC patients and clinicopathological factors. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tau-20-696).
Methods
Data acquisition
Transcriptome profiling data and related clinical information of bladder urothelial carcinoma (BLAC) were downloaded and extracted from The Cancer Genome Atlas (TCGA) Data Portal (https://tcga-data.nci.nih.gov/tcga/; accessed August 2019). Overall, we enrolled a total of 430 BLAC cases containing 411 BC tissue samples and 19 normal bladder tissues into this study. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Identification and enrichment analysis of differently expressed IRGs
We applied “Lima” package in R statistical software to estimate differently expressed IRGs between BC and normal bladder tissues. IRGs exhibiting at least 2-fold changes with an adjusted P value less than 0.05 were identified as the remarkably differentially expressed IRGs. We also performed gene functional enrichment analyses, including Gene Ontology (GO) functional annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, to analyze the biological processes, cellular components, molecular functions and signaling pathways of targeted genes by use of “clusterProfiler” R package.
Identification of differently expressed transcription factors (TFs) and construction of the correlation network between TFs and IRGs
We applied the Cistrome database (http://www.cistrome.org/) to predict TF targets and extract enhancer profiles in cancers. The prediction was from integrative analysis of TCGA expression profiles and public ChIP-seq profiles. Then we identified differently expressed TFs by use of “Lima” package in R statistical software between BLAC and normal bladder tissues. Correlation test between differently expressed TFs and IRGs was conducted by R programming language. Moreover, we set correlation coefficient at least 0.4 with a P value less than 0.01 as the remarkably correlated.
Construction of the PI model based on IRGs
We performed univariate Cox regression analyses to select IRGs significantly associated with OS of BC patients. LASSO-penalized Cox regression model was used to identify the most significantly survival related IRGs. The formula used for calculating the PI is as follow β1 × gene1 expression + β2 ×gene2 expression + ····· + βn × gene expression, where β corresponded to the correlation coefficient.
Evaluation of the PI model
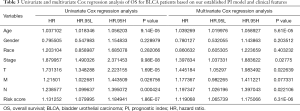
We calculated the risk scores of each patient, and set the median value as the cut-off point to divide the BC patients enrolled into a high-risk group and a low-risk group. Kaplan-Meier plots were generated to analyze the performance of PI model in predicting survival outcomes. The receiver operating characteristic curves (ROC) were created, and the area under the curve (AUC) values was calculated to assess the model specificity and sensitivity. The risk score distribution of patients in different risk groups, and the heatmap of finally confirmed IRGs were also displayed. A prognostic nomogram was also generated to analyze the relationship between individual predictors and survival rates in BC patients. To further determine whether the PI model can be used as an independent prognostic factor, univariate and multivariate cox regression analysis were performed to calculate the HR values for the PI and other clinicopathologic parameters including age, sex, stage, race, grade as well as T, M, N stage. Furthermore, we also conducted a clinical correlation analysis to analyze the correlation between PI and clinicopathologic parameters described above.
Identification of candidate small molecule drugs
Connectivity map (CMap) facilitated researchers to quickly identify molecule drugs highly correlated with diseases and discover its possible mechanism (13). Connectivity scores ranged from −1 to 1 and they were utilized to estimate how closely a compound was connected to the query signature. Negative score indicated that the query signature could be repressed by a drug, while a positive score could be promoted by a drug.
Statistical analysis
We applied R software (version 3.4.1, https://www.r-project.org/) to complete date statistical analyses in this article. It was considered statistically significant when the difference met a joint satisfaction of FDR <0.05 and fold changes ≥2, To establish a prognostic model, univariate and LASSO-penalized COX regression analyses were performed to assess the relationship between IRGs expression and survival data. The receiver operating characteristic curves were created by the “survival ROC” package of R and AUC values were also calculated by this package too. Univariate and multivariate cox regression analyses were to further determine whether our established PI model could be used as an independent prognostic factor. All statistical tests were two-sided and P<0.05 was regarded to be statistically significant.
Results
Differentially expressed IRGs
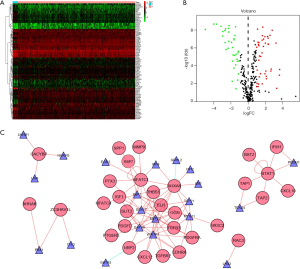
The overall flow diagram for this study was displayed in Figure 1. RNA-seq data of 411 BC tissue samples and 19 normal bladder tissues were downloaded from TCGA. A total of 259 differentially expressed IRGs were extracted with their expression values. The heatmap of differentially expressed IRGs between the BC tissues and normal bladder tissues were demonstrated in Figure 2A. According to the criteria of a FDR <0.05 and |log2(fold change)| >1, we finally obtained 119 up-expressed and 140 down-expressed IRGs (Figure 2B,C).
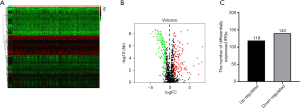
Functional enrichment analysis
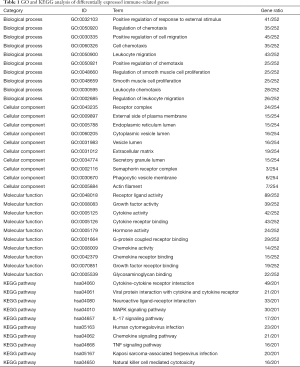
On the basis of the RNA-seq data downloaded from TCGA, we carried out GO term enrichment analysis and KEGG pathway analysis to study the biological function of the differentially expressed IRGs. The GO terms function and KEGG pathway enrichment of these genes were summarized in Table 1.
Full table
As to GO term enrichment analysis, for biological process, the differentially expressed IRGs enriched in positive regulation of response to external stimulus, regulation of chemotaxis, positive regulation of cell migration, cell chemotaxis, leukocyte migration, positive regulation of chemotaxis, regulation of smooth muscle cell proliferation, smooth muscle cell proliferation, leukocyte chemotaxis, regulation of leukocyte migration; for cellular component, the differentially expressed IRGs enriched in receptor complex, external side of plasma membrane, endoplasmic reticulum lumen, cytoplasmic vesicle lumen, vesicle lumen, extracellular matrix, secretory granule lumen, semaphorin receptor complex, phagocytic vesicle membrane, actin filament; For molecular function, the differentially expressed IRGs enriched in receptor ligand activity, growth factor activity, cytokine activity, cytokine receptor binding, hormone activity, G-protein coupled receptor binding, chemokine activity, chemokine receptor binding, growth factor receptor binding, glycosaminoglycan binding (Figure 3A).
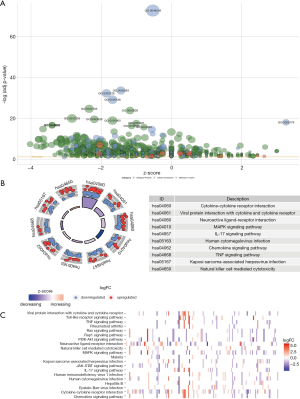
With respect to KEGG pathway analysis, the result showed that the differentially expressed IRGs mainly enriched in cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, neuroactive ligand-receptor interaction, MAPK signaling pathway, IL-17 signaling pathway, and so on (Figure 3B). The Z-score of enriched pathways less than zero indicated that the related pathways were more likely to be decreased. The heatmap of the relationship between IRGs and pathways was also displayed (Figure 3C).
Network analysis of TF-IRG interaction
To explore the interaction between transcription factors (TFs) and IRGs, we extracted 77 differentially expressed TFs by analyzing the RNA-seq data described above. The heatmap of TFs with markedly different expression between the BC tissues and normal bladder tissues were shown in Figure 4A with 41 up-expressed and 36 down-expressed TFs (Figure 4B). The TF-IRG interaction network was detailed in Figure 4C.
Construction of PI model
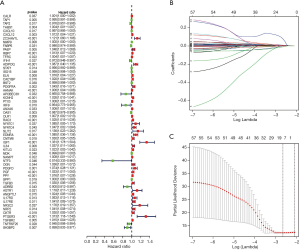
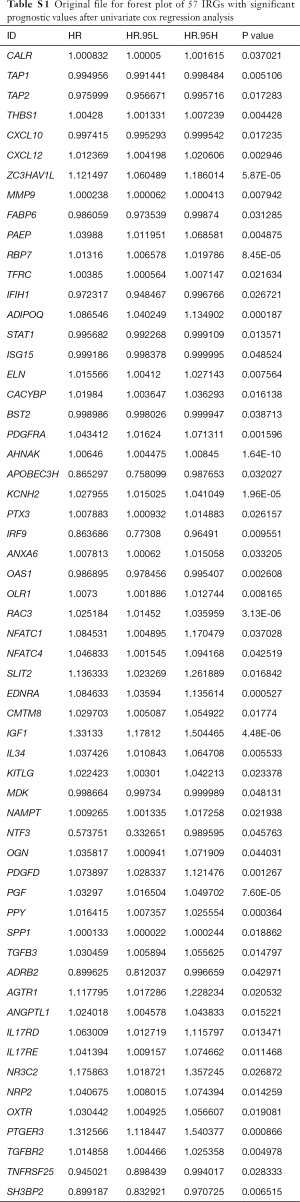
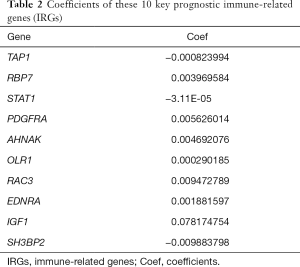
Univariate Cox regression analysis was then performed to investigate the prognostic values of the 259 differentially expressed IRGs. As shown in Figure 5A and Table S1, a total of 57 IRGs were found to be significantly associated with OS in BC patients (P<0.05). Next, LASSO regression analysis was performed to develop the PI model with 10 relevant IRGs (Figure 5B,C). Their specific coefficients of the 10 genes are shown in Table 2. The PI was calculated by use of the formula described above: Risk score = (−0.000824 × TAP1 expression) + (0.003970 ×RBP7 expression) + (−0.000031 × STAT1 expression) + (0.005626 × PDGFRA expression) + (0.004692 ×AHNAK expression) + (0.000290 × OLR1 expression) + (0.009473 × RAC3 expression) + (0.001882 × EDNRA expression) + (0.078175 × IGF1 expression) + (-0.009884 ×SH3BP2 expression).
Full table
Full table
Evaluation of our established PI model
According to the formula described above, we calculated the risk scores of each patient to divide the BC patients into high-risk groups and low-risk groups, based on the median value as the cut-off point. As shown in Figure 6A, with the increase of the risk scores, the patients in high-risk group increased, and the number of dead persons grew. The heatmap of the 10 key prognostic genes expression profiles in the TCGA dataset was also demonstrated. To identify the performance of PI in predicting the clinical outcome of BC patients, the Kaplan-Meier plots were carried out to analyze the different survival time between the high- and low-risk groups. The results showed that patients in the high-risk group had a shorter OS time than patients in the low-risk group (P=9.923e-07, Figure 6B). The ROC curve analysis was performed to evaluate how well the PI predicts the prognoses of BC patients. The AUC for the PI was 0.711, demonstrating the important value of the PI to predict survival of BC patients (Figure 6C). For providing a quantitative approach to predict cancer patient survival, we assembled a nomogram, integrating all the 10 key prognostic genes, which could help estimate 1-, 3-, and 5-year survival probabilities (Figure 6D).
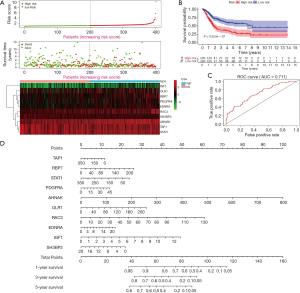
We next performed univariate and multivariate Cox regression analysis to evaluate prognostic value of the PI and other clinical parameters including age, gender, race, tumor stage, T, M, N. The hazard ratios (HRs) for PI in the univariate Cox regression analyses was HR =1.131, 95% confidence interval (CI) =1.080–1.185 (P<0.001, Figure 7A and Table S2), and in the multivariate Cox regression analyses was HR =1.119, 95% CI =1.066–1.175 (P<0.001, Figure 7B and Table S3). As displayed in Figure 7C, it detailed the AUC of all prognostic factors (age, gender, race, tumor stage, T, M, N and risk score) and our established PI model had the highest AUC value. We also integrated both the PI and the clinicopathologic risk factors mentioned above to assemble a nomogram, helping estimate 1-, 3-, and 5-year survival probabilities (Figure 7D). The univariate and multivariate Cox regression analysis of the PI and OS-associated clinical features were shown in Table 3.
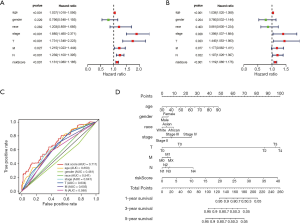
Full table

Full table
Full table
Clinical correlation analysis of the 10 genes and the PI
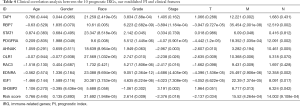
We also performed clinical correlation analysis to study the relationships between the 10 prognostic IRGs, the PI model and the clinicopathologic risk factors. As detailed in Table 4, we found that the PI was positively correlated with race, grade, M and N period (P<0.05), and negatively correlated with stage and T period (P<0.05).
Full table
Related small molecule drugs screening
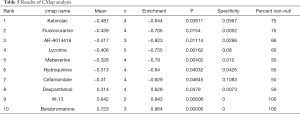
We screened out candidate small molecule drugs of BLAC based on differently expressed IRGs by performing CMap database analysis. By analyzing consistent differently expressed probesets between BLAC samples and adjacent normal samples, we identified the related small molecule drugs with highly significant correlations. 10 small molecule drugs were screened out by the number of instances (n>2) and P value (<0.05). They were shown in Table 5. Among these 10 small molecule drugs, ketorolac, fluorocurarine, AR-A014418, lycorine, mebeverine, hydroquinine, and cefamandole showed negative correlation, and dexpanthenol, W-13, and benzbromarone showed positive correlation. The 7 negatively related small molecules have the potential to treat BLAC, so as the antagonists of the 3 positively related.
Full table
Discussion
As one of the most lethal cancers worldwide (1,14,15), BC has been paid much attention to all the time. In scientific studies, increasing efforts have been put to research the specific mechanisms of BC initiation, progression and prognosis, for the diagnosis, treatment and prognostic surveillance of BC. Many kinds of urinary and blood biomarkers (16-19) were researched for the detection of BC, some of which showed high sensitivity and specificity and may have clinical application value in the future. As reported, long noncoding RNA LNMAT1 and BLACAT2 could promote BC lymphatic metastasis (20,21). And circular RNAs like FNDC3B, ITCH, BCRC-3 and ACVR2A were reported to play important roles in suppressing BC progression (22-25). However, due to high recurrence rate, the long-term efficacy of BC treatment was always unsatisfactory in actual clinical work. Previous research results were not well translated into clinical applications. Hence, it was imperative to develop novel approach to evaluate the prognosis of BC patients, to provide guidance for clinical BC surveillance. In the current study, we concentrated on developing IRGs-based prognostic signature to investigate whether IRG could function as prognostic biomarkers in BC, by analyzing transcriptome data downloaded from TCGA.
In the present study, we first extracted differently-expressed IRGs between BC tissue samples and normal bladder tissues. To identify differently-expressed IRGs with prognostic values, we performed univariate Cox regression analysis and confirmed 57 IRGs. Then, LASSO penalized Cox regression analysis was conducted and 10 IRGs (TAP1, RBP7, STAT1, PDGFRA, AHNAK, OLR1, RAC3, EDNRA, IGF1 and SH3BP2) were identified to develop the PI. Therein, TAP1, a component of the antigen-presenting machinery, was involved in tumor immune escape. It was reported that down-regulation of TAP1 could elicit immune evasion and predict poor prognosis (26,27). Elmasry et al. speculated that in colon cancer, RBP7 functioned as a promotor of tumor migration and invasion through EMT, and indicated poor cancer specific survival (28). As one member of signal transducer and activator of transcription (STAT) family proteins, STAT1 played important roles in tumor microenvironment (29). Meyer Zu Horste et al. demonstrated that STAT1 participated in the process of TH17 differentiation into TH1 (30). PDGFRA mutations were common in gastric, omental and mesenteric tumor (31), and much effort had been made to promote treatments targeting PDGFRA (32-34). AHNAK is a giant structural nucleoprotein involved in diverse biological process including muscle membrane repair, blood-brain barrier, and cell proliferation and migration (35-38). As to OLR1, a lectin-like scavenger receptor, it was mainly expressed in vascular cells and vasculature-rich organs (39), and could function as inflammation promotor in atherogenesis (40). Coincidently, RAC3 could promote ox-LDL induced endothelial dysfunction (41). RAC3 also had correlation with autophagy (41-43), cell migration and invasion (44-46). Much evidence revealed the importance of EDNRA in cardiovascular and cerebrovascular diseases (47-49). As a research hotspot, IGF1 had been receiving much attention and its roles in endocrine disorders, atherosclerosis and cancer promoted development of disease diagnosis and treatment (50-52). SH3BP2 was an adapter protein involved in adaptive and innate immune response signaling, and mainly participated in inflammatory bone loss (53-55). Subsequently, univariate and multivariate Cox regression analysis were carried out to shed light on that this PI model could be an independent predictor. We also analyzed the relationship between this PI model and some clinicopathologic risk factors to confirm their clinical relevance.
Besides, in order to preliminarily explore the relationship between the 57 differentially expressed IRGs and TFs, we constructed a network for them. We first extracted 77 differentially expressed TFs by analyzing the RNA-seq data described above. Then the TF-IRG interaction network was conducted and it could provide a basis for further research. On the one hand, this network analysis was to explore the interactions between TFs and IRGs. On the other hand, it also provided guidance and assistance for further basic researches to seek the upstream regulators of differently expressed IRGs. By analyzing CMap database, we screened out several small molecule drugs with potential therapeutic efficacy to identify candidate small molecule of BLAC. Ketorolac, as a member of non-steroidal anti-inflammatory drugs (NSAIDs) family, was mainly used for inflammation and pain treatment after surgery. In a retrospective analysis of two series of 827 and 1,007 breast cancer patients, Desmedt et al. found that the intra-operative administration of ketorolac was statistically significantly associated with a reduction of distant recurrences (56). AR-A014418, a glycogen synthase kinase-3 inhibitor, could inhibit cell proliferation and induce cell apoptosis in cancers including pancreatic cancer (57) and neuroblastoma (58,59). It also could sensitize pancreatic cancer cells to gemcitabine (60). Wang et al. demonstrated that Lycorine, an alkaloid extracted from Amaryllidaceae genera, could induce apoptosis of BC T24 cells (61). It was reported that anti-calmodulin agent W-13 had the ability to suppress breast cancer cell growth (62) and colony formation (63). As to benzbromarone, it was found to have cytotoxic effects in in human hepatocarcinoma FLC4 cells (64). To our best knowledge, there have no cancer treatment researches of the fluorocurarine, mebeverine, hydroquinine, cefamandole, and dexpanthenol. Overall, our study provided the feasibility of some potential biomarkers or molecular targets in the treatment of BC.
In recent decades, much attention has been paid to researches concentrating on immune in cancer biological and clinical research. T-regulatory (Treg) cells were found to be enriched in BC, and CD4+CD25+ T cells were elevated in patient blood compared with healthy individuals (65). Treg cells were showed to exert the ability to suppress essential antitumor responses (66). Related researches demonstrated that Th17 cells were negatively correlated with Treg cells in BC (67). IL-2 is primarily produced by T-lymphocytes. It was reported that IL-2 could promote the conversion of Treg cells to TH17 cells (68-70). BCG immunotherapy was an important component of BC treatment. Intravesical BCG instillation activated urothelial cells and antigen-presenting cells (APCs), leading to the production of cytokines and chemokines, to further recruit immune cells (71). Cytokines and chemokines like IL-2, IL-6, IL-8, GM-CSF, CC-chemokine ligand 2 (CCL2), and CCL3 could be found in the urine of patients who has underwent BCG treatment (68,72,73). It was reported that IL-2 had correlation with BC recurrence, and along with IL-6, the two cytokines were suggested to be used to monitor BCG treatment effect (74).
The strength of this article was that it was the first time for us to perform a systematic analysis of the roles of immunity in BLCA with a robust statistical approach. A novel PI was successfully established and carefully evaluated. Moreover, networks between IRGs and TFs were also constructed for future basic research. Our study has some limitations. Firstly, we studied only IRGs and other genes associated with OS of BC were not enrolled. Secondly, this study is retrospective and our results should be further validated in prospective investigations.
Conclusions
Taken together, our study successfully developed a novel PI model to predict the OS of BC and it was carefully evaluated. Moreover, networks between TFs and IRGs were also displayed to seek its upstream regulators for future researches. Besides, we also screened out 10 small molecules drugs with the potential to treat BC, based on differently expressed IRGs. Our study highlights the importance of IRGs in clinical application. Although our established PI model stills needs to be further validated by more prospective studies, it was promising to assist clinicians in clinical BC surveillance.
Acknowledgments
We would like to thank the researchers and study participants for their contributions.
Funding: This article was funded by Postdoctoral Science Foundation of Jiangsu Province: 2020Z071.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-696
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-696). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Cumberbatch MGK, Jubber I, Black PC, et al. Epidemiology of Bladder Cancer: A Systematic Review and Contemporary Update of Risk Factors in 2018. Eur Urol 2018;74:784-95. [Crossref] [PubMed]
- Cumberbatch MG, Rota M, Catto JW, et al. The Role of Tobacco Smoke in Bladder and Kidney Carcinogenesis: A Comparison of Exposures and Meta-analysis of Incidence and Mortality Risks. Eur Urol 2016;70:458-66. [Crossref] [PubMed]
- Koutros S, Silverman DT, Alavanja MC, et al. Occupational exposure to pesticides and bladder cancer risk. Int J Epidemiol 2016;45:792-805. [Crossref] [PubMed]
- Fahmy O, Khairul-Asri MG, Schubert T, et al. Urethral recurrence after radical cystectomy for urothelial carcinoma: A systematic review and meta-analysis. Urol Oncol 2018;36:54-9. [Crossref] [PubMed]
- Gakis G, Black PC, Bochner BH, et al. Systematic Review on the Fate of the Remnant Urothelium after Radical Cystectomy. Eur Urol 2017;71:545-57. [Crossref] [PubMed]
- Xu S, Wen Z, Jiang Q, et al. CD58, a novel surface marker, promotes self-renewal of tumor-initiating cells in colorectal cancer. Oncogene 2015;34:1520-31. [Crossref] [PubMed]
- Zhang Y, Zhou X, Zhang M, et al. ZBTB20 promotes cell migration and invasion of gastric cancer by inhibiting IkappaBalpha to induce NF-kappaB activation. Artif Cells Nanomed Biotechnol 2019;47:3862-72. [Crossref] [PubMed]
- Yonemori K, Seki N, Kurahara H, et al. ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci 2017;108:124-35. [Crossref] [PubMed]
- Martínez VG, Rubio C, Martinez-Fernandez M, et al. BMP4 Induces M2 Macrophage Polarization and Favors Tumor Progression in Bladder Cancer. Clin Cancer Res 2017;23:7388-99. [Crossref] [PubMed]
- Ramakrishnan S, Granger V, Rak M, et al. Inhibition of EZH2 induces NK cell-mediated differentiation and death in muscle-invasive bladder cancer. Cell Death Differ 2019;26:2100-14. [Crossref] [PubMed]
- Cheah MT, Chen JY, Sahoo D, et al. CD14-expressing cancer cells establish the inflammatory and proliferative tumor microenvironment in bladder cancer. Proc Natl Acad Sci U S A 2015;112:4725-30. [Crossref] [PubMed]
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929-35. [Crossref] [PubMed]
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Fouad H, Salem H, Ellakwa DE, et al. MMP-2 and MMP-9 as prognostic markers for the early detection of urinary bladder cancer. J Biochem Mol Toxicol 2019;33:e22275. [Crossref] [PubMed]
- Oeyen E, Hoekx L, De Wachter S, et al. Bladder cancer diagnosis and follow-up: the current status and possible role of extracellular vesicles. Int J Mol Sci 2019;20:821. [Crossref] [PubMed]
- Tan WS, Tan WP, Tan MY, et al. Novel urinary biomarkers for the detection of bladder cancer: A systematic review. Cancer Treat Rev 2018;69:39-52. [Crossref] [PubMed]
- Usuba W, Urabe F, Yamamoto Y, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci 2019;110:408-19. [Crossref] [PubMed]
- He W, Zhong G, Jiang N, et al. Long noncoding RNA BLACAT2 promotes bladder cancer-associated lymphangiogenesis and lymphatic metastasis. J Clin Invest 2018;128:861-75. [Crossref] [PubMed]
- Chen C, He W, Huang J, et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat Commun 2018;9:3826. [Crossref] [PubMed]
- Dong W, Bi J, Liu H, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer 2019;18:95. [Crossref] [PubMed]
- Xie F, Li Y, Wang M, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer 2018;17:144. [Crossref] [PubMed]
- Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer 2018;17:19. [Crossref] [PubMed]
- Liu H, Bi J, Dong W, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer 2018;17:161. [Crossref] [PubMed]
- Sultan M, Vidovic D, Paine AS, et al. Epigenetic Silencing of TAP1 in Aldefluor(+) Breast Cancer Stem Cells Contributes to Their Enhanced Immune Evasion. Stem Cells 2018;36:641-54. [Crossref] [PubMed]
- Ling A, Lofgren-Burstrom A, Larsson P, et al. TAP1 down-regulation elicits immune escape and poor prognosis in colorectal cancer. Oncoimmunology 2017;6:e1356143. [Crossref] [PubMed]
- Elmasry M, Brandl L, Engel J, et al. RBP7 is a clinically prognostic biomarker and linked to tumor invasion and EMT in colon cancer. J Cancer 2019;10:4883-91. [Crossref] [PubMed]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009;9:798-809. [Crossref] [PubMed]
- Meyer Zu Horste G, Przybylski D, Schramm MA, et al. Fas Promotes T Helper 17 Cell Differentiation and Inhibits T Helper 1 Cell Development by Binding and Sequestering Transcription Factor STAT1. Immunity 2018;48:556-69.e7. [Crossref] [PubMed]
- Priya V, Kumari N, Krishnani N. Morphological Correlates of KIT and PDGFRA Genotypes in Gastrointestinal Stromal Tumour. Turk Patoloji Derg 2020;36:116-25. [PubMed]
- Kuang FL, Legrand F, Makiya M, et al. Benralizumab for PDGFRA-Negative Hypereosinophilic Syndrome. N Engl J Med 2019;380:1336-46. [Crossref] [PubMed]
- Smith BD, Kaufman MD, Lu WP, et al. Ripretinib (DCC-2618) Is a Switch Control Kinase Inhibitor of a Broad Spectrum of Oncogenic and Drug-Resistant KIT and PDGFRA Variants. Cancer Cell 2019;35:738-51.e9. [Crossref] [PubMed]
- Cools J, DeAngelo DJ, Gotlib J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med 2003;348:1201-14. [Crossref] [PubMed]
- Chen B, Wang J, Dai D, et al. AHNAK suppresses tumour proliferation and invasion by targeting multiple pathways in triple-negative breast cancer. J Exp Clin Cancer Res 2017;36:65. [Crossref] [PubMed]
- Park JW, Kim IY, Choi JW, et al. AHNAK Loss in Mice Promotes Type II Pneumocyte Hyperplasia and Lung Tumor Development. Mol Cancer Res 2018;16:1287-98. [Crossref] [PubMed]
- Sohn M, Shin S, Yoo JY, et al. Ahnak promotes tumor metastasis through transforming growth factor-beta-mediated epithelial-mesenchymal transition. Sci Rep 2018;8:14379. [Crossref] [PubMed]
- Zhang Z, Liu X, Huang R, et al. Upregulation of nucleoprotein AHNAK is associated with poor outcome of pancreatic ductal adenocarcinoma prognosis via mediating epithelial-mesenchymal transition. J Cancer 2019;10:3860-70. [Crossref] [PubMed]
- Yamanaka S, Zhang XY, Miura K, et al. The human gene encoding the lectin-type oxidized LDL receptor (OLR1) is a novel member of the natural killer gene complex with a unique expression profile. Genomics 1998;54:191-9. [Crossref] [PubMed]
- Arslan C, Bayoglu B, Tel C, et al. Upregulation of OLR1 and IL17A genes and their association with blood glucose and lipid levels in femoropopliteal artery disease. Exp Ther Med 2017;13:1160-8. [Crossref] [PubMed]
- He D, Xu L, Wu Y, et al. Rac3, but not Rac1, promotes ox-LDL induced endothelial dysfunction by downregulating autophagy. J Cell Physiol 2020;235:1531-42. [Crossref] [PubMed]
- Xiao X, Wang G, Liu H. Study on the molecular mechanism of Rac3 on regulating autophagy in human lung cancer cells. J BUON 2017;22:445-53. [PubMed]
- Rubio MF, Lira MC, Rosa FD, et al. RAC3 influences the chemoresistance of colon cancer cells through autophagy and apoptosis inhibition. Cancer Cell Int 2017;17:111. [Crossref] [PubMed]
- Zhang C, Liu T, Wang G, et al. Rac3 Regulates Cell Invasion, Migration and EMT in Lung Adenocarcinoma through p38 MAPK Pathway. J Cancer 2017;8:2511-22. [Crossref] [PubMed]
- Donnelly SK, Cabrera R, Mao SPH, et al. Rac3 regulates breast cancer invasion and metastasis by controlling adhesion and matrix degradation. J Cell Biol 2017;216:4331-49. [Crossref] [PubMed]
- Rosenberg BJ, Gil-Henn H, Mader CC, et al. Phosphorylated cortactin recruits Vav2 guanine nucleotide exchange factor to activate Rac3 and promote invadopodial function in invasive breast cancer cells. Mol Biol Cell 2017;28:1347-60. [Crossref] [PubMed]
- Zhang L, Sui R. Effect of SNP polymorphisms of EDN1, EDNRA, and EDNRB gene on ischemic stroke. Cell Biochem Biophys 2014;70:233-9. [Crossref] [PubMed]
- Benjafield AV, Katyk K, Morris BJ. Association of EDNRA, but not WNK4 or FKBP1B, polymorphisms with essential hypertension. Clin Genet 2003;64:433-8. [Crossref] [PubMed]
- Yasuno K, Bakircioglu M, Low SK, et al. Common variant near the endothelin receptor type A (EDNRA) gene is associated with intracranial aneurysm risk. Proc Natl Acad Sci U S A 2011;108:19707-12. [Crossref] [PubMed]
- Clemmons DR. Modifying IGF1 activity: an approach to treat endocrine disorders, atherosclerosis and cancer. Nat Rev Drug Discov 2007;6:821-33. [Crossref] [PubMed]
- Wen HJ, Liu GF, Xiao LZ, et al. Involvement of endothelial nitric oxide synthase pathway in IGF1 protects endothelial progenitor cells against injury from oxidized LDLs. Mol Med Rep 2019;19:660-6. [PubMed]
- Erlandsson MC, Lyngfelt L, Aberg ND, et al. Low serum IGF1 is associated with hypertension and predicts early cardiovascular events in women with rheumatoid arthritis. BMC Med 2019;17:141. [Crossref] [PubMed]
- Mukai T, Fujita S, Morita Y. Tankyrase (PARP5) Inhibition Induces Bone Loss through Accumulation of Its Substrate SH3BP2. Cells 2019;8:195. [Crossref] [PubMed]
- Mukai T, Ishida S, Ishikawa R, et al. SH3BP2 cherubism mutation potentiates TNF-alpha-induced osteoclastogenesis via NFATc1 and TNF-alpha-mediated inflammatory bone loss. J Bone Miner Res 2014;29:2618-35. [Crossref] [PubMed]
- Reichenberger EJ, Levine MA, Olsen BR, et al. The role of SH3BP2 in the pathophysiology of cherubism. Orphanet J Rare Dis 2012;7 Suppl 1:S5. [Crossref] [PubMed]
- Desmedt C, Demicheli R, Fornili M, et al. Potential Benefit of Intra-operative Administration of Ketorolac on Breast Cancer Recurrence According to the Patient's Body Mass Index. J Natl Cancer Inst 2018;110:1115-22. [Crossref] [PubMed]
- Kunnimalaiyaan S, Gamblin TC, Kunnimalaiyaan M. Glycogen synthase kinase-3 inhibitor AR-A014418 suppresses pancreatic cancer cell growth via inhibition of GSK-3-mediated Notch1 expression. HPB (Oxford) 2015;17:770-6. [Crossref] [PubMed]
- Carter YM, Kunnimalaiyaan S, Chen H, et al. Specific glycogen synthase kinase-3 inhibition reduces neuroendocrine markers and suppresses neuroblastoma cell growth. Cancer Biol Ther 2014;15:510-5. [Crossref] [PubMed]
- Dent P. New methods to control neuroblastoma growth. Cancer Biol Ther 2014;15:481-2. [Crossref] [PubMed]
- Shimasaki T, Ishigaki Y, Nakamura Y, et al. Glycogen synthase kinase 3beta inhibition sensitizes pancreatic cancer cells to gemcitabine. J Gastroenterol 2012;47:321-33. [Crossref] [PubMed]
- Wang C, Wang Q, Li X, et al. Lycorine induces apoptosis of bladder cancer T24 cells by inhibiting phospho-Akt and activating the intrinsic apoptotic cascade. Biochem Biophys Res Commun 2017;483:197-202. [Crossref] [PubMed]
- Strobl JS, Peterson VA. Tamoxifen-resistant human breast cancer cell growth: inhibition by thioridazine, pimozide and the calmodulin antagonist, W-13. J Pharmacol Exp Ther 1992;263:186-93. [PubMed]
- Wei JW, Hickie RA, Klaassen DJ. Inhibition of human breast cancer colony formation by anticalmodulin agents: trifluoperazine, W-7, and W-13. Cancer Chemother Pharmacol 1983;11:86-90. [Crossref] [PubMed]
- Kobayashi K, Kajiwara E, Ishikawa M, et al. Cytotoxic effects of benzbromarone and its 1'-hydroxy metabolite in human hepatocarcinoma FLC4 cells cultured on micro-space cell culture plates. Drug Metab Pharmacokinet 2013;28:265-8. [Crossref] [PubMed]
- Loskog A, Ninalga C, Paul-Wetterberg G, et al. Human bladder carcinoma is dominated by T-regulatory cells and Th1 inhibitory cytokines. J Urol 2007;177:353-8. [Crossref] [PubMed]
- Nishikawa H, Sakaguchi S. Regulatory T cells in tumor immunity. Int J Cancer 2010;127:759-67. [PubMed]
- Chi LJ, Lu HT, Li GL, et al. Involvement of T helper type 17 and regulatory T cell activity in tumour immunology of bladder carcinoma. Clin Exp Immunol 2010;161:480-9. [Crossref] [PubMed]
- Thompson DB, Siref LE, Feloney MP, et al. Immunological basis in the pathogenesis and treatment of bladder cancer. Expert Rev Clin Immunol 2015;11:265-79. [Crossref] [PubMed]
- Dudley ME, Wunderlich JR, Yang JC, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol 2005;23:2346-57. [Crossref] [PubMed]
- Kryczek I, Wei S, Zou L, et al. Cutting edge: Th17 and regulatory T cell dynamics and the regulation by IL-2 in the tumor microenvironment. J Immunol 2007;178:6730-3. [Crossref] [PubMed]
- Redelman-Sidi G, Glickman MS, Bochner BH. The mechanism of action of BCG therapy for bladder cancer--a current perspective. Nat Rev Urol 2014;11:153-62. [Crossref] [PubMed]
- De Boer EC, De Jong WH, Steerenberg PA, et al. Induction of urinary interleukin-1 (IL-1), IL-2, IL-6, and tumour necrosis factor during intravesical immunotherapy with bacillus Calmette-Guerin in superficial bladder cancer. Cancer Immunol Immunother 1992;34:306-12. [Crossref] [PubMed]
- Böhle A, Brandau S. Immune mechanisms in bacillus Calmette-Guerin immunotherapy for superficial bladder cancer. J Urol 2003;170:964-9. [Crossref] [PubMed]
- de Reijke TM, de Boer EC, Kurth KH, et al. Urinary cytokines during intravesical bacillus Calmette-Guerin therapy for superficial bladder cancer: processing, stability and prognostic value. J Urol 1996;155:477-82. [Crossref] [PubMed]