
Identification of an independent autophagy-gene prognostic index for papillary renal cell carcinoma
Introduction
Renal cell carcinoma (RCC), as the most common malignancy in renal neoplasia, caused nearly 12,000 deaths annually worldwide (1). Papillary RCC, accounting for 15% in RCC, was one of the common subtypes of RCC. With lower histological grade and distant organ metastasis, papillary RCC was generally considered to have better disease-free survival (DFS) and overall survival (OS) than clear RCC. Recommended therapy for papillary included partial nephrectomy, nephrectomy, radiofrequency ablation, molecular target therapy and administration of immune checkpoint inhibitor (2). Despite of great progress with regard to therapy and pathogenesis of papillary achieved recent years, prognosis of papillary RCC was still worse. So, the exploration of underlying molecular mechanisms of tumorigenesis and metastasis in papillary RCC was still urgent nowadays.
Autophagy, a highly conserved and evolutionarily ancient biological process in yeast and eukaryotic cells, played a significant role in maintaining the homeostasis of cell in stressed states. The fundamental process of autophagy contained the activation of PI3KC3 complex and initiation of isolated membrane, elongation and closure of autophagosome membrane and formation of autolysosome (3,4). The dysregulation of autophagy was now found accounted for many pathological neurodegenerative diseases (5), cardiomyopathy (6). Some researched have pointed out the momentous role of autophagy in the tumorigenesis and metastasis in papillary RCC (7-9). Besides, the administration of chloroquine (CQ), an inhibitor of formation of autophagosomes, have been found to increase the survival of patients and have synergetic effects when combining with other molecular target drugs (10). However, the concrete role of autophagy in RCC still remained controversial and mysterious.
In this study, we tried to illustrate a comprehensive relation between autophagy and papillary RCC. At first, we identified 40 autophagy related genes (ARGs) which differently expressed genes between adjacent tumor tissues and tumor tissues. Then, by applying COX regression model, 4 core ARGs (P4HB, BIRC5, NGR1 and PRKN) were selected for the further analysis. Finally, we developed prognostic index (PI) based on these 4 core ARGs as an independent index for predicting the OS in papillary RCC and clinical parameters were extracted to reveal the relationship between these 4 core ARGs and clinical status. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-906).
Materials
Data extraction
We identified 234 ARGs from The Human Autophagy Database (HADb, http://www.autophagy.lu/index.html), an autophagy-related database which recorded ARGs that have been described in literature. Gene profile of level 3 RNA-sequence in The Cancer Genome Atlas (TCGA) cohort was downloaded by using R Studio and only data regarding papillary RCC was extracted for further analysis by checking pathological examination. Then, some clinical parameters like gender, Stage, pathological grade, TMN stage, vital status and survival time were also acquired.
Differently expressed gene enrichment analysis
By using EdgeR package in R Studio, 40 differently expressed ARGs were estimated by comparing tumor tissues and adjacent tumor tissues, with the cut-off criterion of fold change >2 and P value <0.01. Then, in order to explore the major biological process of these 40 ARGs, these genes were utilized to conduct the gene functional enrichment analyses including Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO). We utilized Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) to identify enriched KEGG and GO themes.
Establishment of a PI based on ARGs
In order to find out ARGs which gene expression were associated with OS in patients with papillary RCC, several genes were identified by using univariate Cox regression analyses. Then, we conducted multivariate Cox regression analysis to obtain genes that might be an independent indicator. Totally, 4 ARGs (P4HB, BIRC5, NGR1 and PRKN) were selected for the construction of PI. The construction of PI based on a linear combination of the relative expression level of genes multiplied regression coefficients. The risk coefficients of ARGs were calculated by utilizing a multivariable Cox proportional hazards model. According to the median PI value as the risk cutoff value, all the patients with papillary RCC were divided into low- and high- risk groups. OS of these two groups were analyzed by Kaplan-Meier method and the difference between low and high groups were tested by log-rank test. Finally, we plotted receiver operating characteristic curves (ROCs) to verify the prediction value of the model.
Statistical analysis
The relationship between clinical parameters and gene expression profile was analyzed by Fisher’s exact test or chi-square test. Continuous variables were compared by Student’s t-test. The OS analyses were performed by the Kaplan-Meier method and log-rank test. We carried out all statistical analyses using R Studio 3.6.1 (https://www.r-project.org/), SPSS 24.0 (Chicago, IL, USA) and GraphPad Prism 8 (San Diego, CA, USA). Probability P value <0.05 was considered to be statistically significant.
Ethical statement
All data from TCGA are publicly available and their use do not require the approval of a local ethics committee. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Validation of ARGs in papillary RCC
Totally, after checking the pathological examination of all the RCC tissues, 289 papillary RCC tissues and 32 adjacent tumor tissues were downloaded from TCGA (Table 1). Among them, 45 patients with gene expression profile and clinical information were included for the further analysis. We compared the differently expressed genes between tumor tissues and adjacent tumor tissues by applying EdgeR package in R Studio. With P value <0.01 and fold change >2 as the cut-off criteria, 31 up-regulated ARGs and 9 down-regulated ARGs were obtained in the end. Volcano map, heat map and box plots were carried out to visualize the differently expressed ARGs (Figure 1).
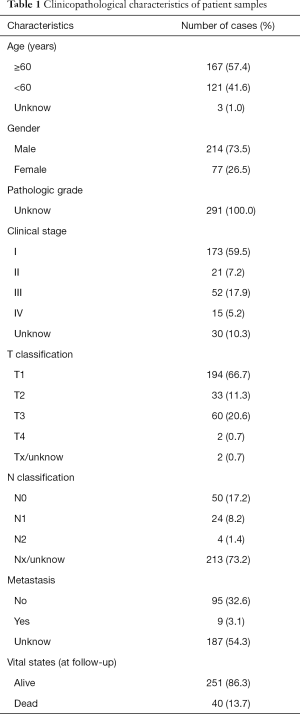
Full table
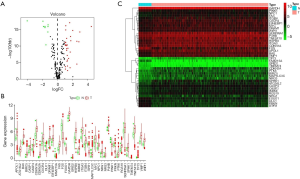
Functional annotation of these 40 differentially expressed ARGs
In order to fully understand the biological attributes of these 40 differentially expressed ARGs, we conducted KEGG and GO analysis. Based on the results of DAVID, the top ten enriched GO terms were: selective autophagy, response to toxic response, response to starvation, response to oxygen level, regulation of release of cytochrome c from mitochondria, regulation of peptidase activity, regulation of autophagy, process utilizing autophagic mechanism, regulation of endopeptidase activity and positive regulation of tumor necrosis factor-mediated signaling pathway (Figure 2A,B,C). The top biological pathway enriched were: human cytomegalovirus infection, hepatitis B, Kaposi sarcoma-associated herpesvirus infection (Figure 2D,E,F).
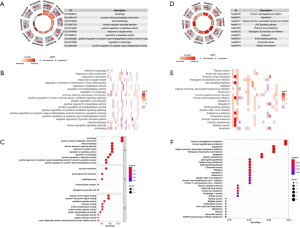
Identification of prognostic ARGs and construction of the PI
For the purpose of revealing the relationship between these 40 ARGs and OS, 12 prognostic ARGs were identified by utilizing univariate Cox regression analyses (Figure 3A and Table 1). Then, multivariate Cox regression analysis was applied to increase the robustness and select the independent indicators for the OS and finally we got 4 ARGs (P4HB, BIRC5, NGR1 and PRKN) for the further analysis [P4HB: hazard ratio (HR) =2.686, 95% confidence interval (CI): 1.394–5.172, P=0.003; BIRC5: HR =2.290, 95% CI: 1.604–3.272, P<0.001; PRKN: HR =0.502, 95% CI: 0.255–0.986, P=0.046; NGR1: HR =1.652, 95% CI: 1.095–2.494, P=0.017] (Table 2). After the construction of PI, patients were separated into high- and low- risk (Figure 3B). Heap map was utilized to visualize the difference of gene expression profile in low- and high- risk patients in papillary RCC (Figure 3C). The results from Kaplan-Meier analysis indicated that high-risk patients had lower OS than low-risk patients in papillary RCC (P<0.001) (Figure 3D). Further, we applied this prognostic model to predict the vital status of patients and we found that the mortality of high scoring patients was higher than low scoring patients (Figure 3E).
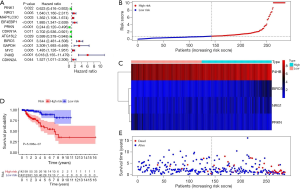

Full table
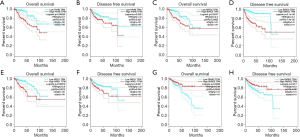
Based on the Kaplan-Meier analysis, we found that patients with higher P4HB expression level showed an inferior OS and DFS (HR =3.1, P=0.00055, Figure 4A; HR =1.9, P=0.029, Figure 4B). Also, overexpression BIRC5 and NRG1 indicated worse OS and DFS (HR =2.4, P=0.0047, Figure 4C; HR =4.4, P=2.1e-06, Figure 4D; HR =2.9, P=0.00072, Figure 4E; HR =1.8, P=0.041, Figure 4F). On the contrary, patients with higher PARK2 expression had a better DFS and OS (HR =0.39, P=0.0029, Figure 4G; HR =0.5, P=0.017, Figure 4H). This might reveal that PARK2 was positive to better clinical outcomes whereas P4HB, BIRC5, NRG1 were negative.
Relationship between 4 key genes and clinical parameters
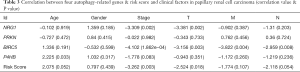
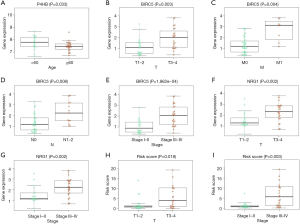
We tried to illustrate the association between some clinical parameters and the gene expression level of these 4 key ARGs (P4HB, BIRC5, NGR1 and PRKN) (Table 3). Clinical parameters including T stage, N stage, M stage, age, gender and Stage were extracted in the current study. We found that the expression level of P4HB was correlated with age of patients. Patients <60 years old had higher P4HB expression level than patients >60 years old (P=0.033) (Figure 5A). As for BIRC5, its gene expression level was associated with T stage, M stage, N stage and Stage (P=0.003, P=0.004, P=0.008, P=1.862e-04, respectively) (Figure 5B,C,D,E). Patients with higher stage (T3–4, M1, N1–2, Stage III–IV) had higher BIRC5 expression than patients with lower stage (T1–2, N0, M0, Stage I–II). NRG1 was found to have relationship with T stage and Stage (P=0.002, Figure 5F; P=0.002, Figure 5G). Again, patients with higher NRG1 expression level showed a higher T stage and Stage (T3–4, Stage III–IV). Besides, no clinical parameters were illustrated to correlate with PRKN. Finally, we found that high scoring patients had higher T stage and Stage (T3–4, Stage III–IV) than low scoring patients (T1–2, Stage I–II) while other clinical parameters showed no significance (P=0.018, Figure 5H; P=0.003, Figure 5I).
Full table
The PI was an independent prognostic factor for papillary RCC
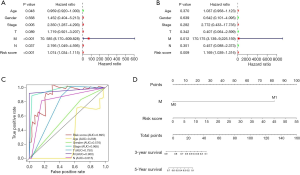
In order to investigate whether the risk signature was independent from clinical parameters such as patients’ age, gender, Stage, lymphatic invasion, tumor size and metastasis, we conducted a univariate Cox and multivariate Cox regression analysis and explored the area under curve (AUC) (Figure 6). According to the results, we found that metastasis stage and risk-score were significantly correlated with OS. Finally, we came to the conclusion that our risk signature was an independent prognostic factors for papillary RCC. To further provide a more quantitative and comprehensive prediction for the 3- and 5-year survival probability, we integrated out risk signature and some clinicopathological factors to construct a nomogram (Figure 6D).
Discussion
While RCC was one of the most common malignancy in renal neoplasia, papillary RCC was a significant subtype of it. Thus, it was urgent to find molecular biomarkers that could aid in a better prognosis and OS for papillary RCC patients. It seemed that exploration of autophagy mechanism had shed light on this new perspective. To date, numerous studies have focused on the role that autophagy played in tumorigenesis and prognosis. Although autophagy has been a hotspot, it still remained a controversy whether autophagy suppressed or promoted tumorigenesis and metastasis. For example, several pathways such as p53 status, RAS family status, activation of JAK-STAT and PI3K signaling might all impose influence on the determination of autophagy dependence within cancer cells (11). Researches have shown that autophagy suppressed the initiation of tumor while promoting the progression of tumor (12). This implied that ARGs could act as a potential anticancer target in the therapeutic strategies and the prognosis of patients. However, while many researches of autophagy-related genes relied on the data retrieved from cell lines or animal models, our current study took use of high-throughput data of ARGs to investigate the prognosis and clinical outcomes of patients with papillary RCC. What’s more, we also explored the relationship between 4 key genes and clinical parameters, which could contribute to a better understanding of our prognostic signature.
Given the great development of high-throughput sequencing and the emergence of large-scare databases, we were able to select gene signatures. First, to investigate the differentially expressed ARGs, we compared the different expression level of autophagy between tumor tissues and adjacent tumor tissues and obtained 40 autophagy-related genes, including some popular oncotherapeutic target genes such as BIRC5 and NRG1. In order to figure out the functional attributes of these 40 ARGs, we then performed GO and KEGG analysis to figure out the biological attributes of these genes. Results demonstrated that these enriched genes were often associated with the following pathways: p53 signaling pathway, apoptosis, cellular senescence and so on. Relationship between p53 and autophagy has been observed. It seemed that autophagy could promote cancer by repressing p53, which was an important mechanism of tumorigenesis. What’s more, p53 also activated the transcription of ARGs and thus promoted autophagy (13). The functional association between autophagy and apoptosis was rather complex. Frequently, autophagy could serve as a trigger of apoptosis and aided in the process of the cellular suicide (14). For cellular senescence, however, it remained in controversy whether autophagy influenced senescence positively or negatively. Kang et al. claimed that selective autophagy functioned as an antisenescence mechanism, whereas general autophagy functioned as a pro-senescence mechanism (15).
Based on the results of univariate Cox regression analysis, 12 ARGs associated with OS were picked out. To further increase the robustness, multivariate Cox regression analysis was performed and finally 4 independent prognostic ARGs (P4HB, BIRC5, NGR1 and PRKN) were screen out to develop the PI. After constructing the 4-gene signature, patients were separated into high-risk and low-risk groups. Our results showed that high risk-score patients had an inferior survival probability and clinical outcomes.
BIRC5 (also named as survivin) was a well-known cancer therapeutic target which inhibited apoptosis and regulated mitosis both in embryonic cells during embryogenesis and in cancer cells during tumorigenesis (16). Studies have found that the mammalian target of rapamycin (mTOR) pathway was one of important pathways of autophagy, the inhibition of which was correlated with the promotion of autophagy (17). The post-transcription regulation of BIRC5 might be mediated by mTOR to increase the mRNA stability and translation of it (18). This might reveal the relationship between autophagy and BIRC5. Besides, the overexpression of BIRC5 was observed in almost all human malignancies, and the increased expression of BIRC5 was correlated with poor clinical outcomes, tumour recurrence and drug resistance in cancer patients (19). Our results showed that enriched gene expression of BIRC5 was associated with worse clinical index, which meant the coefficient of BIRC5 in the PI was positive and was in accordance with the accepted opinion of BIRC5 function in cancer. P4HB was a multifunctional protein which encoded the beta subunit of prolyl 4-hydroxylase and catalyzed the formation and rearrangement of disulfide bonds. Zhou et al. found that knockdown of P4HB could inhibit proliferation and promote apoptosis of human HT29 colon cancer cells via accumulation of reactive oxygen species and inhibition of STAT3 signaling (20). Several studies have reported that P4HB was significantly increased in several solid tumors including bladder cancer, brain and central nervous system (CNS) cancer, lung cancer, prostate cancer (21) and was correlated with poor prognosis. Xie et al. and Zhu et al. had shown that P4HB overexpressed in human clear cell RCC and was correlated with a poor prognosis, confirming that P4HB might be a novel biomarker (21,22). According to our results, P4HB showed a higher expression level in samples under 60 years old than those over 60 (P=0.03). Researches demonstrated that aging was accompanied by reduced autophagy level in various organisms, which was in accordance with our results. This phenomenon might result from an aging-dependent reduced expression of genes important for autophagosome-lysosome fusion, such as lysosome-associated membrane protein 2 (LAMP2A) (23). NRG1, a member of the NRG family, was a ligand for human epidermal growth factor (HER) 3 and HER4 which could activate cell signaling pathways to promote tumorigenesis and metastasis (24). Studies have indicated that NRG1 was overexpressed in various cancer such as breast cancer and gastric cancer (24,25). Han et al. found that increased NRG1 expression was related to advanced pathological stage, lymphnode metastasis and poor prognosis (26). Our research showed that expression of NRG was higher in T3–4 stage than in T1–2 stage, which was in accordance with Han et al. This indicated that NRG was associated with unfavourable clinicopathologic features in papillary RCC and was a negative prognostic factor. Research that targeted NRG1 pathway in lung cancer has succeeded, which meant NRG1 was an independent and effective biomarker for treatment and provided a novel strategy (27).
The advantage of our study was that instead of using cell lines and animal model, we turned to high-throughput data and large-scale databases to conduct a 4-gene prognostic signature statistically, which catered to the urgent need of an effective index for papillary RCC. Besides, our study could provide a better understanding of the role of autophagy in papillary RCC. However, our study did have some limitations. First of all, clinical parameters such as age, pathological stage were not integrated into our PI formula. We mainly focused on molecular markers while clinical perspective should also be explored. Second, our risk signature was composed of only ARGs and did not represent other potential gene transcription expression profiles correlated to OS in papillary RCC.
To date, the use of prognostic signature was proposed due to rapid development of RNAseq. Our study developed an independent 4-gene signature to reflect the clinical outcomes of papillary RCC. Further work will be done to deeply illustrate the role of autophagy in tumor and to improve our prognostic signature. In all, this novel therapy could provide a promising future for both diagnosis and prognosis for not only papillary RCC but also various cancer.
Acknowledgments
Funding: This work was supported by the Jiangsu Province Six Talent Peaks Project (WSN-011), and the National Natural Science Foundation of China (grant number 81672531, 81972386).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-906
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-906). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All data from TCGA are publicly available and their use do not require the approval of a local ethics committee. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. [Crossref] [PubMed]
- Ljungberg B, Bensalah K, Canfield S, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol 2015;67:913-24. [Crossref] [PubMed]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000;290:1717-21. [Crossref] [PubMed]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011;147:728-41. [Crossref] [PubMed]
- Tan CC, Yu JT, Tan MS, et al. Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol Aging 2014;35:941-57. [Crossref] [PubMed]
- Taneike M, Yamaguchi O, Nakai A, et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 2010;6:600-6. [Crossref] [PubMed]
- Li H, Jin X, Zhang Z, et al. Inhibition of autophagy enhances apoptosis induced by the PI3K/AKT/mTor inhibitor NVP-BEZ235 in renal cell carcinoma cells. Cell Biochem Funct 2013;31:427-33. [Crossref] [PubMed]
- Zhang Y, Fan Y, Huang S, et al. Thymoquinone inhibits the metastasis of renal cell cancer cells by inducing autophagy via AMPK/mTOR signaling pathway. Cancer Sci 2018;109:3865-73. [Crossref] [PubMed]
- Zheng B, Zhu H, Gu D, et al. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun 2015;459:234-9. [Crossref] [PubMed]
- Li ML, Xu YZ, Lu WJ, et al. Chloroquine potentiates the anticancer effect of sunitinib on renal cell carcinoma by inhibiting autophagy and inducing apoptosis. Oncol Lett 2018;15:2839-46. [PubMed]
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17:528-42. [Crossref] [PubMed]
- Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev 2016;30:1913-30. [Crossref] [PubMed]
- White E. Autophagy and p53. Cold Spring Harb Perspect Med 2016;6:a026120. [Crossref] [PubMed]
- Maiuri MC, Zalckvar E, Kimchi A, et al. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007;8:741-52. [Crossref] [PubMed]
- Kang C, Elledge SJ. How autophagy both activates and inhibits cellular senescence. Autophagy 2016;12:898-9. [Crossref] [PubMed]
- Lin TY, Chan HH, Chen SH, et al. BIRC5/Survivin is a novel ATG12-ATG5 conjugate interactor and an autophagy-induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy 2020;16:1296-313. [Crossref] [PubMed]
- Cao Q, Bai P. Role of Autophagy in Renal Cancer. J Cancer 2019;10:2501-9. [Crossref] [PubMed]
- Roca H, Varsos ZS, Mizutani K, et al. CCL2, survivin and autophagy: new links with implications in human cancer. Autophagy 2008;4:969-71. [Crossref] [PubMed]
- Lyu H, Wang S, Huang J, et al. Survivin-targeting miR-542-3p overcomes HER3 signaling-induced chemoresistance and enhances the antitumor activity of paclitaxel against HER2-overexpressing breast cancer. Cancer Lett 2018;420:97-108. [Crossref] [PubMed]
- Zhou Y, Yang J, Zhang Q, et al. P4HB knockdown induces human HT29 colon cancer cell apoptosis through the generation of reactive oxygen species and inactivation of STAT3 signaling. Mol Med Rep 2019;19:231-7. [PubMed]
- Xie L, Li H, Zhang L, et al. Autophagy-related gene P4HB: a novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging (Albany NY) 2020;12:1828-42. [Crossref] [PubMed]
- Zhu Z, He A, Lv T, et al. Overexpression of P4HB is correlated with poor prognosis in human clear cell renal cell carcinoma. Cancer Biomark 2019;26:431-9. [Crossref] [PubMed]
- Leidal AM, Levine B, Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol 2018;20:1338-48. [Crossref] [PubMed]
- Yun S, Koh J, Nam SK, et al. Clinical significance of overexpression of NRG1 and its receptors, HER3 and HER4, in gastric cancer patients. Gastric Cancer 2018;21:225-36. [Crossref] [PubMed]
- Yang L, Li Y, Shen E, et al. NRG1-dependent activation of HER3 induces primary resistance to trastuzumab in HER2-overexpressing breast cancer cells. Int J Oncol 2017;51:1553-62. [Crossref] [PubMed]
- Han ME, Kim HJ, Shin DH, et al. Overexpression of NRG1 promotes progression of gastric cancer by regulating the self-renewal of cancer stem cells. J Gastroenterol 2015;50:645-56. [Crossref] [PubMed]
- Jones MR, Lim H, Shen Y, et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer. Ann Oncol 2017;28:3092-7. [Crossref] [PubMed]


