
Construction of a risk signature for adrenocortical carcinoma using immune-related genes
Introduction
Adrenocortical carcinoma (ACC), which originates from the cortex of the adrenal gland, is considered a rare tumor with a low incidence rate of 2 per million people (1). However, among endocrine carcinomas, the aggressiveness of ACC is only lower than that of anaplastic thyroid cancer, and patients with ACC at an advanced clinical stage have a dismal prognosis. The median overall survival (OS) time and 5-year survival rate of ACC patients are 3.21 years and 15% to 44%, respectively (2-5). At present, the only effective treatment for ACC is radical resection, which is recommended only when the size of the tumor exceeds 5 cm or increased circulating adrenal hormone levels are confirmed (6). Adjuvant therapies, such as mitoxantrone combined with radiotherapy, administered after surgery can delay ACC recurrence (7).
Recent studies have indicated that a disrupted immune system, the main factor that allows tumor cells to evade the immune response, is involved in the development and progression of tumors (8). As a promising antitumor strategy, immunotherapy aims to induce the immune system to recognize cancer antigens as foreign antigens and suppress the proliferation and metastasis of tumor cells through active and passive immune responses, thus accelerating the development of personalized medicine (9,10). Many studies have suggested a relationship between ACC and immunity. For example, one study showed that among the three ACC subsets integrated through cluster analysis, immune-mediated pathways were significantly upregulated in the subset with the best prognosis (11). Loss of major histocompatibility complex, class II, DR beta 1 (HLA-DRB1), a major histocompatibility complex (MHC) class II allele, and altered expression of Fas/Fas ligand are possible mechanisms of immune escape in ACC (6). A novel adjuvant for immunotherapy, autologous dendritic cells can induce tumor-specific responses in ACC patients, even though final outcomes have not been satisfactory (12). High levels of CD8+ T lymphocytes can independently improve prognosis in childhood ACC (13). Furthermore, upregulation of the immune markers neutrophil-lymphocyte ratio (NLR) and platelet-lymphocyte ratio (PLR) correlates significantly with a poor prognosis in ACC patients after resection (14). Although these findings show that immunity is important in ACC, the associated molecular mechanisms, particularly those underlying immunogenomic effects, remain unclear. Hence, we assessed expression of immune-related genes (IRGs) in ACC and investigated correlations between IRGs and ACC prognosis using public datasets. Gene expression and clinical data for ACC patients were downloaded from The Cancer Genome Atlas (TCGA), and gene expression data for the normal adrenal gland were obtained from the Genotype-Tissue Expression (GTEx) database. The authors present the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tau-20-485).
Methods
Characteristics of the included ACC patients
Our study was a retrospective analysis. Transcriptomic data and corresponding clinical data for ACC patients were downloaded from GTEx and TCGA in the UCSC Xena database (http://xena.ucsc.edu/). We normalized and merged the transcriptome data from 79 ACC tissues and 127 normal adrenal tissues from the two databases into a single dataset to identify differentially expressed genes (DEGs). Seventy-seven ACC patients with both gene expression data and corresponding clinical data were included for additional analysis. OS was used as a prognostic indicator, and the median follow-up time was 1,171 days. Data for the 77 ACC patients are shown in Table 1. IRG information was selected from the Immunology Database and Analysis Portal (ImmPort) database (https://www.immport.org/) (15). To further explore regulatory mechanisms, we also obtained transcription factor (TF) information from the Cistrome Cancer database (http://cistrome.org/) (16). All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and no ethical approval was required because the data we used were obtained from public databases. Because of the retrospective nature of the research, the requirement for informed consent was waived.
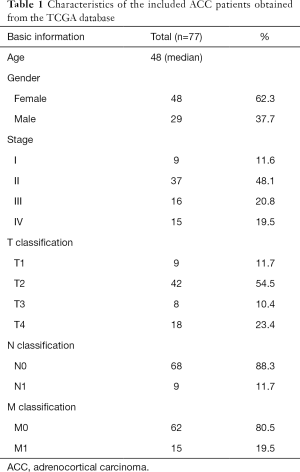
Full table
Immune score
An immune score was obtained using the transcriptome data of ACC tissues in TCGA via Estimation of Stromal and Immune cells in Malignant Tumor tissues with Expression data (ESTIMATE), an algorithm that calculates immune and stromal scores based on expression of immune cells and stromal cell-related genes in tumors to predict infiltration of these cells. The current study only focused on the immune score (17,18). Then, we generated Kaplan-Meier survival curves to illustrate the relationship between ACC patient OS and the immune score (the median value of the immune score was used as the cutoff value).
DEGs
We compared expression of genes in 79 ACC tissue samples with that in 127 normal tissue samples via the edgeR package in R software (19). |Log2 fold change (FC)| >1.0 and a false discovery rate (FDR) adjusted to P<0.05 were defined as the cutoff criteria. Differentially expressed IRGs and TFs were identified from the DEGs. We carried out Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to estimate the biological functions of the differentially expressed IRGs initially via the clusterProfiler package in R software (20) and visualized these results using the GOplot package in R software (21).
Molecular mechanisms of IRGs associated with survival
We performed univariate Cox regression to extract IRGs associated with survival, setting a P value <0.01 as the cutoff. Protein-protein interaction (PPI) network analysis with the String database (score >0.4) (https://stringdb.org/) was conducted to assess relationships among the IRGs associated with survival in ACC patients. In the PPI network, each node represents the protein product of the IRGs associated with survival and each edge the interaction between the proteins (22). Genes with the largest number of edges, also called hub genes, were distinguished in our PPI network. Moreover, as regulatory mechanisms were the focus of the current study, we constructed a TF regulatory network (P value <0.05 and correlation coefficient >0.4 defined as the cutoff criteria) using Cytoscape software version 3.7.2 for visualization.
Risk signature
IRGs associated with survival were subjected to multivariate Cox regression to determine the coefficient of each selected IRG, and the risk score was calculated based on the formula below.
We divided patients into two groups (the median value of the risk score was used as the cutoff value): high-risk and low-risk groups. A receiver operating characteristic (ROC) curve was built to evaluate the accuracy of the risk score in predicting the prognosis of ACC patients (24). Fisher’s exact tests were applied to explore differences in clinical variables between the groups. Kaplan-Meier survival curves and Cox regression were utilized to confirm that the risk score was able to independently predict OS. The clinical variables included for evaluating the effectiveness of the signature were age, gender, TNM classification and clinical stage.
Statistical analysis
The Wilcoxon rank-sum test was used for comparisons between two groups, whereas the Kruskal-Wallis test with Bonferroni’s post hoc test was performed for multigroup comparisons. The survival curve represented by the Kaplan-Meier survival curve was analyzed for the difference in OS between the two groups via the log-rank test. Cox regression was conducted by using the survival package in R software.
Results
Immune score
We initially found that compared with a low score, a high immune score predicted a better prognosis and early clinical-stage disease in ACC, indicating that immunotherapy may be effective for ACC treatment (Figure 1A,B). Relatively high immune scores were also observed in male patients and patients with advanced TNM-stage disease, but there were no significant differences (Figure 1C,D,E,F). These results showed that a high level of immune cell infiltration can affect the prognosis of ACC patients and is related to some early clinical variables.
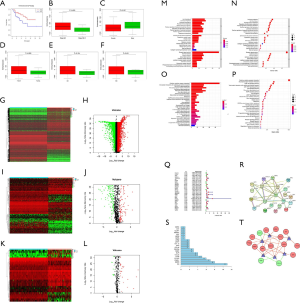
DEGs in ACC
We identified 5,045 DEGs from all genes in the transcriptomic dataset obtained from UCSC Xena; 2,646 of the DEGs were upregulated and 2,399 downregulated (Figure 1G,H). Of the 5,045 DEGs, 227 differentially expressed IRGs were screened, with 93 exhibiting upregulated expression and 134 exhibiting downregulated expression (Figure 1I,J). Finally, we identified 33 differentially expressed TFs, which consisted of 22 with upregulated expression and 11 with downregulated expression (Figure 1K,L). The results of GO and KEGG analyses for the differentially expressed IRGs are illustrated in Figure 1M,N,O,P. According to the GO results, receptor ligand activity was the most frequent GO biological process category. The “cytokine-cytokine receptor interaction” was identified as the most enriched pathway in KEGG analyses.
IRGs associated with survival
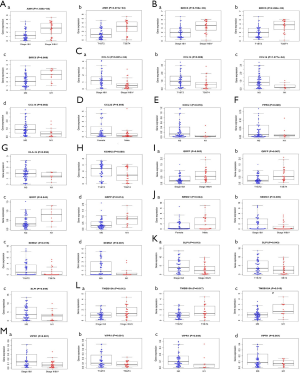
We extracted 30 IRGs significantly associated with survival via univariate Cox regression (all P values <0.01) (Figure 1Q). A PPI network that included all 30 IRGs associated with survival was established, with scores of >0.4 in String (Figure 1R). In the PPI network, interleukin 6 (IL-6) contained the largest number of edges; thus, it was identified as the hub gene of the IRGs associated with survival in ACC (Figure 1S). To further investigate associated regulatory mechanisms, a TF regulatory network was constructed based on the 33 differentially expressed TFs and 30 IRGs associated with survival (Figure 1T). In addition, we evaluated correlations between clinicopathological factors and IRGs associated with survival (Figure 2).
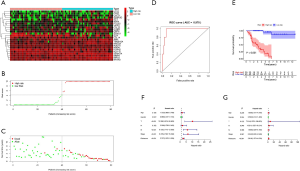
High risk scores indicated a poor prognosis in ACC
The risk score was calculated from coefficients determined by multivariate Cox regression. A risk signature was then constructed, dividing the patients into two groups (the median value of the risk score was used as the cutoff value), namely, a high-risk group and a low-risk group (Figure 3A,B,C). The coefficients of the included genes obtained from multivariate Cox regression are presented in Table 2.
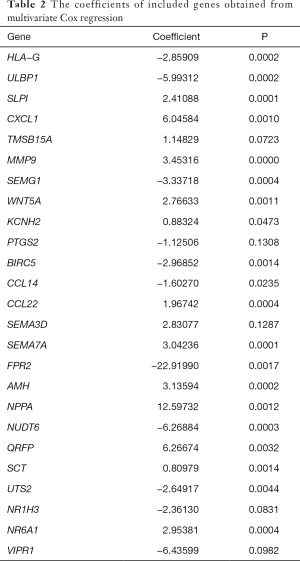
Full table
The area under the ROC curve (AUC) was 0.970, suggesting that the risk signature had the potential to accurately predict the prognosis of ACC patients (Figure 3D). The results of Kaplan-Meier survival analyses and Fisher’s exact tests revealed that a high risk score correlated significantly with an advanced clinical stage (P=0.011), a high T classification (P=0.004), a high M classification (P=0.010) and poor OS (P<0.001) (Figure 3E and Table 3). Univariate Cox regression demonstrated that T classification [hazard ratio (HR) =10.286; 95% confidence interval (CI): 3.976–26.608; P<0.001), M classification (HR =6.150; 95% CI: 2.710–13.959; P<0.001), clinical stage (HR =6.476; 95% CI: 2.706–15.498; P<0.001) and the risk score (HR =2.707; 95% CI: 2.070–3.539; P<0.001) could predict prognosis (Figure 3F, Table 3). We then carried out multivariate Cox regression, which showed that the risk score (HR =2.819; 95% CI: 1.973–4.030; P<0.001) was associated with prognosis (Figure 3G). Therefore, we considered the risk score to be an independent prognostic factor for ACC.
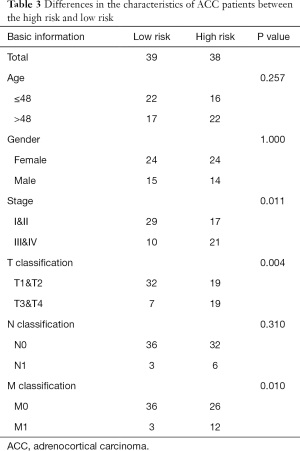
Full table
Discussion
Currently, immunotherapy, which exploits the immune system to fight tumors, is a potential treatment for various types of malignancies. Nevertheless, new findings suggest that tumor cells can escape the immune response by using immune checkpoints, such as programmed death-1 (PD-1), programmed death ligand-1 (PD-L1) and cytotoxic T lymphocyte antigen-4 (CTLA-4), in the tumor microenvironment (25). Therefore, research on the application of immune checkpoint inhibitors to prevent immune escape is receiving much attention at present (26-28). In ACC, PD-L1 expression is detected on the membrane of tumor cells, and silencing of mismatch repair genes, which can improve sensitivity to anti-PD-1 therapy, has been demonstrated in 30% of ACC patients, suggesting that immunotherapy may be an effective treatment for ACC (29-31). A recent study showed that IFNγ activates immune cell infiltration in anti-ACC therapy but that it might increase expression of PD-L1, suppressing the immune response. Although it ultimately improved the postoperative disease-free survival (DFS) of ACC patients, the effect of the immune response was not satisfactory. Therefore, the application of anti-PD-L1 therapy might alleviate its suppression of the immune response (32). In addition to expression of immune checkpoint molecules, reduced signaling and expression of the innate immune receptor Toll-like receptor 4 (TLR4) was recently reported in ACC, providing a novel marker for immunotherapy in ACC (33). Although immunotherapy provides hope for the treatment of ACC, there are still some challenges. For instance, immune cytolytic activity in ACC is lower than that in other cancers, and immune responses are inhibited by the secretion of steroid hormones, an important feature of ACC; both of these are considered major barriers to effective immunotherapy in ACC (25,34). Unfortunately, research on immunology and ACC is limited, and the scarcity of case samples caused by the low incidence of ACC may be the main reason. Therefore, further analyses of IRGs using transcriptomic data from public databases will be important for the prediction of ACC prognosis and the development of immunotherapeutic strategies.
In this study, we initially identified that compared with a low score, a high immune score predicted a better prognosis and early clinical stage disease in ACC, indicating that immunotherapy may be effective for ACC treatment. Differentially expressed IRGs and TFs were then extracted, and KEGG functional enrichment analysis showed that the five most significant signaling pathways for activation of the differentially expressed IRGs were the PI3K-Akt signaling pathway, JAK-STAT signaling pathway, chemokine signaling pathway, Ras signaling pathway and MAPK signaling pathway. Previous studies have shown that ACC cell proliferation is partially activated by PI3K-Akt and MAPK signaling (35-37). Mutations in Ras-related genes have also been observed in ACC, and we found that the JAK-STAT signaling promoted ACC cell aldosterone secretion (38-42). Studies on the specific mechanisms underlying chemokine signaling and the relationships between these five signaling pathways and immunotherapy in ACC have rarely been reported. Thus, we propose for the first time that these pathways are associated with immunotherapy efficacy in ACC.
IRGs associated with survival were screened by univariate Cox regression. IL-6 was confirmed to be the hub gene in the PPI network. Among the IRGs associated with survival in ACC patients, a higher level of IL-6 was detected in ACC samples than in normal samples; a high IL-6 level can stimulate the secretion of glucocorticoids, and the serum level of matrix metallopeptidase 9 (MMP-9) decreases significantly after radical resection (43-45). Then, we constructed a TF regulatory network based on IRGs associated with survival and differentially expressed TFs. Among all differentially expressed TFs in ACC patients, centromere protein A (CENPA) (46), E2F transcription factor 1 (E2F1) (47) and forkhead box M1 (FOMX1) (48) showed upregulated expression that was involved in ACC progression and predicted a poor prognosis. In contrast, downregulation of transcription factor 21 (TCF21) expression resulted in the accumulation of secreted glucocorticoids and accelerated proliferation of ACC cells, indicating that TCF21 is a potential prognostic marker in ACC (49-51). Additionally, a previous study reported that Activin could induce x-zone apoptosis to inhibit ACC cell growth induced by SMAD family member 2 (Smad2) (52). However, no studies on other IRGs associated with survival or TFs in ACC are available, and previous publications only offer limited information about the regulatory mechanisms involved. Hence, we constructed a TF regulatory network for further analysis. Conclusively, a risk signature was built from the coefficients of selected IRGs determined by multivariate Cox regression, and it was validated that the risk score can serve as an independent prognostic factor for ACC.
Nonetheless, the deficiencies in our study should be acknowledged. First, we merged transcriptomic data from two databases due to the small number of ACC samples in TCGA and the lack of paired normal tissue samples. Second, our analysis of IRG functions was not verified by in vitro or in vivo experiments. Third, the infiltration of each specific immune cell type was not estimated clearly.
In conclusion, we assessed the roles of IRGs in ACC and investigated correlations between IRGs and ACC prognosis. The signature based on IRGs may be used as a tool to predict prognosis in patients with ACC, and our results provide preliminary evidence for the application of immunotherapy in ACC. Although our findings offer a novel perspective for immunotherapy in ACC, further studies are needed.
Acknowledgments
We are also grateful for the data provided by TCGA and GTEx for this study.
Funding: The authors are grateful to the Project of Liaoning Distinguished Professor (Grant No. [2012]145), the Shenyang Plan Project of Science and Technology (Grant No. F17-230-9-08), China Medical University’s 2017 Discipline Promotion Program (Grant No. 3110117040), China Medical University’s 2018 Discipline Promotion Program, and the 2017 National Key R&D Program Key Projects of Precision Medical Research (No. 2017YFC0908000).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-485
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-485). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and no ethical approval was required because the data we used were obtained from public databases. Because of the retrospective nature of the research, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bilimoria KY, Shen WT, Elaraj D, et al. Adrenocortical carcinoma in the United States: treatment utilization and prognostic factors. Cancer 2008;113:3130-6. [Crossref] [PubMed]
- Schteingart DE, Doherty GM, Gauger PG, et al. Management of patients with adrenal cancer: recommendations of an international consensus conference. Endocr Relat Cancer 2005;12:667-80. [Crossref] [PubMed]
- Xiao H, He W, Chen P, et al. Identification of Seven Aberrantly Methylated and Expressed Genes in Adrenocortical Carcinoma. Front Endocrinol (Lausanne) 2019;10:472. [Crossref] [PubMed]
- Tella SH, Kommalapati A, Yaturu S, et al. Predictors of Survival in Adrenocortical Carcinoma: An Analysis From the National Cancer Database. J Clin Endocrinol Metab 2018;103:3566-73. [Crossref] [PubMed]
- Ranvier GG, Inabnet WB 3rd. Surgical management of adrenocortical carcinoma. Endocrinol Metab Clin North Am 2015;44:435-52. [Crossref] [PubMed]
- Wolkersdorfer GW, Marx C, Brown J, et al. Prevalence of HLA-DRB1 genotype and altered Fas/Fas ligand expression in adrenocortical carcinoma. J Clin Endocrinol Metab 2005;90:1768-74. [Crossref] [PubMed]
- Else T, Williams AR, Sabolch A, et al. Adjuvant therapies and patient and tumor characteristics associated with survival of adult patients with adrenocortical carcinoma. J Clin Endocrinol Metab 2014;99:455-61. [Crossref] [PubMed]
- Duan S, Wang P, Liu F, et al. Novel immune-risk score of gastric cancer: A molecular prediction model combining the value of immune-risk status and chemosensitivity. Cancer Med 2019;8:2675-85. [Crossref] [PubMed]
- Carter BW, Halpenny DF, Ginsberg MS, et al. Immunotherapy in Non-Small Cell Lung Cancer Treatment: Current Status and the Role of Imaging. J Thorac Imaging 2017;32:300-12. [Crossref] [PubMed]
- Lin P, Guo YN, Shi L, et al. Development of a prognostic index based on an immunogenomic landscape analysis of papillary thyroid cancer. Aging (Albany NY) 2019;11:480-500. [Crossref] [PubMed]
- Zheng S, Cherniack AD, Dewal N, et al. Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma. Cancer Cell 2016;29:723-36. [Crossref] [PubMed]
- Papewalis C, Fassnacht M, Willenberg HS, et al. Dendritic cells as potential adjuvant for immunotherapy in adrenocortical carcinoma. Clin Endocrinol (Oxf) 2006;65:215-22. [Crossref] [PubMed]
- Parise IZS, Parise GA, Noronha L, et al. The Prognostic Role of CD8(+) T Lymphocytes in Childhood Adrenocortical Carcinomas Compared to Ki-67, PD-1, PD-L1, and the Weiss Score. Cancers (Basel) 2019;11:1730. [Crossref] [PubMed]
- Bagante F, Tran TB, Postlewait LM, et al. Neutrophil-lymphocyte and platelet-lymphocyte ratio as predictors of disease specific survival after resection of adrenocortical carcinoma. J Surg Oncol 2015;112:164-72. [Crossref] [PubMed]
- Bhattacharya S, Andorf S, Gomes L, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res 2014;58:234-9. [Crossref] [PubMed]
- Mei S, Meyer CA, Zheng R, et al. Cistrome Cancer: A Web Resource for Integrative Gene Regulation Modeling in Cancer. Cancer Res 2017;77:e19-22. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Li B, Geng R, Wu Q, et al. Alterations in Immune-Related Genes as Potential Marker of Prognosis in Breast Cancer. Front Oncol 2020;10:333. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284-7. [Crossref] [PubMed]
- Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015;31:2912-4. [Crossref] [PubMed]
- Zhang Z, Chen D, Li Z, et al. Bioinformatics Analysis to Screen the Key Prognostic Genes in Tumor Microenvironment of Bladder Cancer. Biomed Res Int 2020;2020:6034670. [Crossref] [PubMed]
- Chai RC, Wu F, Wang QX, et al. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY) 2019;11:1204-25. [Crossref] [PubMed]
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000;56:337-44. [Crossref] [PubMed]
- Roufas C, Chasiotis D, Makris A, et al. The Expression and Prognostic Impact of Immune Cytolytic Activity-Related Markers in Human Malignancies: A Comprehensive Meta-analysis. Front Oncol 2018;8:27. [Crossref] [PubMed]
- Cunha LL, Marcello MA, Rocha-Santos V, et al. Immunotherapy against endocrine malignancies: immune checkpoint inhibitors lead the way. Endocr Relat Cancer 2017;24:T261-81. [Crossref] [PubMed]
- Popovic A, Jaffee EM, Zaidi N. Emerging strategies for combination checkpoint modulators in cancer immunotherapy. J Clin Invest 2018;128:3209-18. [Crossref] [PubMed]
- Kobold S, Pantelyushin S, Rataj F, et al. Rationale for Combining Bispecific T Cell Activating Antibodies With Checkpoint Blockade for Cancer Therapy. Front Oncol 2018;8:285. [Crossref] [PubMed]
- Mohan DR, Lerario AM, Hammer GD. Therapeutic Targets for Adrenocortical Carcinoma in the Genomics Era. J Endocr Soc 2018;2:1259-74. [Crossref] [PubMed]
- Fay AP, Signoretti S, Callea M, et al. Programmed death ligand-1 expression in adrenocortical carcinoma: an exploratory biomarker study. J Immunother Cancer 2015;3:3. [Crossref] [PubMed]
- Mandal R, Samstein RM, Lee KW, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 2019;364:485-91. [Crossref] [PubMed]
- Billon E, Finetti P, Bertucci A, et al. PDL1 expression is associated with longer postoperative, survival in adrenocortical carcinoma. Oncoimmunology 2019;8:e1655362. [Crossref] [PubMed]
- Kanczkowski W, Tymoszuk P, Ehrhart-Bornstein M, et al. Abrogation of TLR4 and CD14 expression and signaling in human adrenocortical tumors. J Clin Endocrinol Metab 2010;95:E421-9. [Crossref] [PubMed]
- Nakamura Y, Yamazaki Y, Felizola SJ, et al. Adrenocortical carcinoma: review of the pathologic features, production of adrenal steroids, and molecular pathogenesis. Endocrinol Metab Clin North Am 2015;44:399-410. [Crossref] [PubMed]
- Lee M, Waser B, Reubi JC, et al. Secretin receptor promotes the proliferation of endocrine tumor cells via the PI3K/AKT pathway. Mol Endocrinol 2012;26:1394-405. [Crossref] [PubMed]
- Pezzani R, Rubin B, Redaelli M, et al. The antiproliferative effects of ouabain and everolimus on adrenocortical tumor cells. Endocr J 2014;61:41-53. [Crossref] [PubMed]
- Andreis PG, Rucinski M, Neri G, et al. Neuropeptides B and W enhance the growth of human adrenocortical carcinoma-derived NCI-H295 cells by exerting MAPK p42/p44-mediated proliferogenic and antiapoptotic effects. Int J Mol Med 2005;16:1021-8. [Crossref] [PubMed]
- Kotoula V, Sozopoulos E, Litsiou H, et al. Mutational analysis of the BRAF, RAS and EGFR genes in human adrenocortical carcinomas. Endocr Relat Cancer 2009;16:565-72. [Crossref] [PubMed]
- Kirschner LS. Signaling pathways in adrenocortical cancer. Ann N Y Acad Sci 2002;968:222-39. [Crossref] [PubMed]
- Costa R, Wesolowski R, Raghavan D. Chemotherapy for advanced adrenal cancer: improvement from a molecular approach? BJU Int 2011;108:1546-54. [Crossref] [PubMed]
- Yashiro T, Hara H, Fulton NC, et al. Point mutations of ras genes in human adrenal cortical tumors: absence in adrenocortical hyperplasia. World J Surg 1994;18:455-60; discussion 460-1. [Crossref] [PubMed]
- Itcho K, Oki K, Kobuke K, et al. Angiotensin 1-7 suppresses angiotensin II mediated aldosterone production via JAK/STAT signaling inhibition. J Steroid Biochem Mol Biol 2019;185:137-41. [Crossref] [PubMed]
- Spath-Schwalbe E, Born J, Schrezenmeier H, et al. Interleukin-6 stimulates the hypothalamus-pituitary-adrenocortical axis in man. J Clin Endocrinol Metab 1994;79:1212-4. [PubMed]
- Babinska A, Kaszubowski M, Kmiec P, et al. Adipokine and cytokine levels in patients with adrenocortical cancer, subclinical Cushing’s syndrome and healthy controls. Steroids 2018;140:39-44. [Crossref] [PubMed]
- Kolomecki K, Stepien H, Bartos M, et al. Evaluation of MMP-1, MMP-8, MMP-9 serum levels in patients with adrenal tumors prior to and after surgery. Neoplasma 2001;48:116-21. [PubMed]
- Xia WX, Yu Q, Li GH, et al. Identification of four hub genes associated with adrenocortical carcinoma progression by WGCNA. PeerJ 2019;7:e6555. [Crossref] [PubMed]
- Tabbal H, Septier A, Mathieu M, et al. EZH2 cooperates with E2F1 to stimulate expression of genes involved in adrenocortical carcinoma aggressiveness. Br J Cancer 2019;121:384-94. [Crossref] [PubMed]
- Kar A, Zhang Y, Yacob B, et al. Targeting PDZ binding kinase is anti-tumorigenic in novel preclinical models of ACC. Endocr Relat Cancer 2019;26:765-78. [Crossref] [PubMed]
- Franca MM, Lerario AM, Fragoso M, et al. New evidences on the regulation of SF-1 expression by POD1/TCF21 in adrenocortical tumor cells. Clinics (Sao Paulo) 2017;72:391-4. [Crossref] [PubMed]
- Franca MM, Ferraz-de-Souza B, Santos MG, et al. POD-1 binding to the E-box sequence inhibits SF-1 and StAR expression in human adrenocortical tumor cells. Mol Cell Endocrinol 2013;371:140-7. [Crossref] [PubMed]
- Passaia BDS, Dias MH, Kremer JL, et al. TCF21/POD-1, a Transcritional Regulator of SF-1/NR5A1, as a Potential Prognosis Marker in Adult and Pediatric Adrenocortical Tumors. Front Endocrinol (Lausanne) 2018;9:38. [Crossref] [PubMed]
- Beuschlein F, Looyenga BD, Bleasdale SE, et al. Activin induces x-zone apoptosis that inhibits luteinizing hormone-dependent adrenocortical tumor formation in inhibin-deficient mice. Mol Cell Biol 2003;23:3951-64. [Crossref] [PubMed]


