The level of zinc finger of the cerebellum 2 is predictive of overall survival in clear cell renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) encompasses cancer originated from renal tubular epithelial cells and clear cell renal cell carcinoma (ccRCC) accounts for the majority of RCC cases (1). In 2017, an estimated 64,000 patients were diagnosed with, and 14,400 patients died from, ccRCC worldwide (2). Though most patients experienced slow-growing, non-lethal tumors, approximately 20–40% of ccRCC patients may suffer from recurrence and metastasis after curative nephrectomy during the follow-up period (3,4). Moreover, current clinical and pathologic variables alone are insufficient to distinguish indolent and aggressive tumors and predict prognosis for most patients. In addition, identifying factors that could predict oncologic outcomes is important in many ways, such as patient counseling, and healthcare policy development (3).
Zinc finger of the cerebellum 2 (ZIC2) belongs to a family of five genes originally identified due to their homology to the drosophila odd-paired genes. ZIC2 may play a major role in the growing and maturing of central nervous system, because heterozygous deletions or mutations of ZIC2 could lead to severe brain malformation (5,6). Intriguingly, ZIC2 was also found to be highly expressed in some tumors and involved in oncogenesis (7-10). ZIC2 was overexpressed in epithelial ovarian, non-small cell lung, endometrial, hepatocellular, prostate and cervical cancers and correlated with poor clinical outcomes (10-12). For example, Marchini et al. reported that increased ZIC2 was significantly related to the clinical features of patients with epithelial ovarian carcinoma (7). Furthermore, ZIC2 has also been confirmed to be functionally significant at the molecular and cellular levels in the formation and progression of prostate carcinoma (12). Increased ZIC2 expression also facilitated hepatocellular cancer growth and metastasis via up-regulating the P21-Activated kinase 4 (PAK4) in hepatocellular carcinoma, and promoted cell proliferation by retaining GlI1 in the nucleus of cervical cancer (11,13).
Bioinformatic analysis aims to group together data based on biological similarities accessing different sources of research, allowing different types of data accessed for researchers in the process of data analysis (14). The Cancer Genome Atlas (TCGA) is widely used in bioinformatic analysis because it makes molecular profile data available to the scientific community for performing further analyses of data from multi-institutional databases (15). Using TCGA several genes, such as nitrogen permease regulator-like 2 (NPRL2), perilipin 3 (PLIN3), chemokine (C-X-C motif) receptor 4 (CXCR4), and discoidin domain receptor tyrosine kinase 1 (DDR1) have been found to be associated with the diagnosis or prognosis of ccRCC by bioinformatic analysis (16), however, the level of ZIC2 expression and its relationship with oncologic outcomes have not been analyzed previously in ccRCC patients. Considering the biological and clinical importance of ZIC2 and its oncogenic features (7), we aimed to investigate the prognostic significance of ZIC2 by bioinformatics analysis in ccRCC patients.
Methods
Bioinformatic analysis using UALCAN
UALCAN is an interactive web-portal, which is adopted to compare the difference in the level of genes between tumor samples and normal kidney tissues and associations between survival and tumor clinicopathological features (http://ualcan.path.uab.edu/analysis.html) (17). The expression level of the ZIC2 mRNA in ccRCC samples compared to normal kidney tissues was analyzed through UALCAN. UALCAN analysis was employed to explore the impacts of ZIC2 expression according to tumor-node-metastasis (TNM) stages and histologic grades. The data used in this study is available freely from TCGA database (https://xenabrowser.net/).
Original data obtained with the help of UCSC Xena browser
The original data of ccRCC patients was obtained from TCGA, with the help of the University of California Santa Cruz Xena browser (https://xenabrowser.net/) (18). Then the associations between the level of ZIC2 and clinicopathological features, such as age, T, N, M, TNM stage and histologic grade, were investigated. Furthermore, we explored its association with overall survival (OS) and disease-free survival (DFS) and prognostic value in ccRCC patients.
Statistical analysis
Statistical analyses employed a SPSS statistical software package 22.0 (IBM, Armonk, NY, USA). The statistical difference between the level of ZIC2 in ccRCC and normal samples was analyzed by t-test. The associations between the level of ZIC2 and clinicopathological characteristics were assessed by χ2 tests. The appropriate cut-off point value for the level of ZIC2 was confirmed by receiver operating characteristic curve analyses with the maximal value of Youden index. OS and DFS were evaluated by Kaplan–Meier analyses and log-rank test was conducted to assess differences between groups. Multivariate Cox regression analyses were adopted to investigate the significance of ZIC2 in OS and DFS. Two-sided P<0.05 was of statistical significance.
Results
ZIC2 is significantly elevated in ccRCC compared with normal samples
By using online analyses tool UALCAN, we identified the expression of ZIC2 mRNA in 533 ccRCC and 72 normal kidney samples. Results revealed the level of ZIC2 was significantly higher in ccRCC than that in normal kidney samples (Figure 1A). Significantly different expression levels were also found by different TNM stages and histologic grades. ZIC2 expression increased with the escalation of TNM stages and histologic grades (Figure 1B,C).

Associations between ZIC2 mRNA expression and clinicopathological characteristics
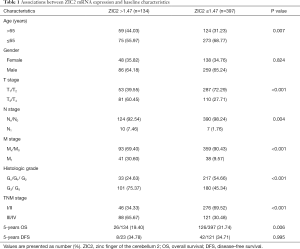
Table 1 showed the correlations between the level of ZIC2 and baseline characteristics in ccRCC patients. Patients in the high level of ZIC2 group were with advanced age (age >65), T, N, M, TNM stage, and histologic grade than those in the low one. In addition, a significantly lower 5-years OS (19.40% vs. 31.74%, P=0.006) was also found in the patients with the higher level of ZIC2 compared with the other group, whereas no significant differences in 5-years DFS between the two groups were noted.
Full table
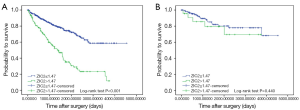
ZIC2 mRNA expression is related to significantly worse OS in ccRCC patients
High ZIC2 expression is associated with significantly worse OS (P<0.001) in ccRCC patients (Figure 2A). However, no statistical correlation was found between the expression of ZIC2 and DFS, which may due to the small number of patients included (Figure 2B).
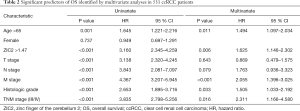
Elevated ZIC2 expression is an independent predictor for worse OS in ccRCC patients
Univariate analyses indicated that advanced TNM stage (III/IV), age (age>65), M stage, histologic grade and increased level of ZIC2 were correlated with dramatically worse OS in ccRCC patients. Unsurprisingly, multivariate analyses demonstrated that the elevated level of ZIC2 was an independent predictor for worse OS (HR: 1.625, 95% CI, 1.146–2.302, P=0.006) (Table 2).
Full table
Discussion
To the best of our knowledge, this study is the first to examine the value of ZIC2 in the prediction of survival in ccRCC patients. In this study, we found that the increased level of ZIC2 was correlated with significantly worse OS and an independent prognostic factor in ccRCC patients, though no statistically significant association was found between the level of ZIC2 expression and DFS, which may due to the small number of patients included.
In human cancers, the dysregulation of transcription factors (TFs) is usually implicated in the pathogenesis (11). ZIC2 is a member of the five ZIC genes which encode C2H2-type zinc finger TFs. Notwithstanding ZIC2 was only reported in the brain and testis in normal tissues, up-regulated ZIC2 has been found in cancers of the lung, endometrium, bladder, pancreas, cervix, breast and prostate, as well as, specifically pediatric medulloblastoma, synovial sarcoma, oral squamous cell carcinoma, nasopharyngeal carcinoma, and hepatocellular carcinoma (8,9,12,13,19-23). Interestingly, ZIC2 was downregulated in pediatric medulloblastoma, which could be explained by the methylation of the CpG islands (24). However, biometric analysis of the level of ZIC2 expression in ccRCC patients had never been reported before. This study demonstrated that the level of ZIC2 was significantly higher in ccRCC samples than that in normal ones and increased with the escalation of TNM stages and histologic grades.
The associations between ZIC2 overexpression and the clinical course and prognosis of patients with tumors were also reported in previous studies. Specifically, ZIC2 mRNA was significantly higher expressed in endometrial cancers with positive nodes compared with those with negative nodes, and in the malignant form of epithelial ovarian tumors compared with those with low malignant potential (7,20). Moreover, high expression of ZIC2 was correlated with a worse outcome in patients with stage I epithelial ovarian cancer, cervical cancer, oral squamous cell and hepatocellular carcinoma. Levels of ZIC2 were an independent indicator of poor outcome (7,11,13,25). The same was true in ccRCC patients. Though no statistical relation was detected between the level of ZIC2 and DFS, the high level of ZIC2 was correlated with the worse OS in ccRCC patients, which may be attributed to its association with advanced age, T, N, M, TNM stage, and histologic grade to a certain extent.
Several molecular mechanisms involved in the oncogenic properties of ZIC2 have been studied to some extent. ZIC2 was demonstrated to be a positive modulator of GLI1 protein, with GLI1 retained in the nucleus, thereby, enhancing its transcriptional and oncogenic activity as well as activation of Hedgehog signalling. For example, ZIC2 up-regulation promoted cell proliferation in cervical cancer by enhancing Hedgehog signaling (13). Another report showed that overexpression of ZIC2 could markedly induce cell proliferation and migration via the Raf/MEK/ERK pathway through transcriptionally up-regulating PAK4 in hepatocellular carcinoma (11). High expression of ZIC2 enhanced the cellular proliferation of pancreatic ductal adenocarcinoma and suppressed cell apoptosis by up-regulating fibroblast growth factor receptor 3 and ANNEXIN A8, which is independent of the levels of GLI1 and GLI2 (23). Bawa et al. reported that ZIC2 was recruited to the ABCC4 promoter region by lncRNA prostate cancer associated transcript 92 and subsequently up-regulate the level of ATP-binding cassette sub-family C member 4 (12). In breast cancer cells, ZIC2 was targeted by microRNA-1284 to regulate cell proliferation, migration, invasion and apoptosis through PI3K/AKT pathway (8). Taken together, ZIC2 functions as an oncogenic transcription factor, making ZIC2 as a new target for therapy in various types of tumors.
However, little is known about the mechanisms underlying ZIC2-overexpression in ccRCC patients, which still needs further study undoubtedly. Expectedly, recent studies suggested that the levels of GLI1 and GLI2 expression were significantly elevated in a large cohort of ccRCC patients, which were negatively correlated with patient OS (26,27). According to the above results, it is reasonable to suppose that over-expression of ZIC2 may up-regulate the expression of GLI1 and active the Hedgehog signaling, promoting oncogenesis or tumor progression accordingly. In addition, it should be also noted that unlike other prognostic indicators correlated with only some of the tumor grades or pathological features, the level of ZIC2 was associated to both, which may indicate the important role of ZIC2 in ccRCC progression. Interestingly, a significant correlation was found between renal/urinary defects and mutations of ZIC2 (28).
Considering some limitations, the results of our study should be interpreted cautiously. Firstly, the retrospective design of this study with data acquired from TCGA database inevitably results in considerable bias. Thus, larger, multicenter and prospective studies are needed to verify our findings and the diagnostic value of ZIC2 combined with NPRL2, PLIN3, CXCR4 to ccRCC patients. Secondly, analyzed by different detection methods and platforms, the level of ZIC2 expression may be inaccurate and of inter-study heterogeneity due to the lack of uniform standardization. Moreover, the small sample size of present research may also limit the reliability and stability of our conclusions. Finally, neither the specific function of ZIC2 in ccRCC nor molecular mechanisms implicated in that were explored by in vitro or in vivo experiments.
Conclusions
In summary, our bioinformatics analysis suggests the indispensable and particular effects of ZIC2 in the survival of ccRCC patients.
Acknowledgments
The results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.Z Shang would like to thank Miss Xiaolin Liu for her valuable understanding and assistance.
Funding: This work was supported by the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (grant number ZYLX201801).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.01.12). The authors have no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Xuanwu Hospital Capital Medical University (No. 2019-23).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers 2017;3:17009. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Leibovich BC, Lohse CM, Cheville JC, et al. Predicting Oncologic Outcomes in Renal Cell Carcinoma After Surgery. Eur Urol 2018;73:772-80. [Crossref] [PubMed]
- Sun M, Marconi L, Eisen T, et al. Adjuvant Vascular Endothelial Growth Factor-targeted Therapy in Renal Cell Carcinoma: A Systematic Review and Pooled Analysis. Eur Urol 2018;74:611-20. [Crossref] [PubMed]
- Zhu P, Wang Y, He L, et al. ZIC2-dependent OCT4 activation drives self-renewal of human liver cancer stem cells. J Clin Invest 2015;125:3795-808. [Crossref] [PubMed]
- García-Frigola C, Carreres MI, Vegar C, et al. Zic2 promotes axonal divergence at the optic chiasm midline by EphB1-dependent and -independent mechanisms. Development 2008;135:1833-41. [Crossref] [PubMed]
- Marchini S, Poynor E, Barakat RR, et al. The zinc finger gene ZIC2 has features of an oncogene and its overexpression correlates strongly with the clinical course of epithelial ovarian cancer. Clin Cancer Res 2012;18:4313-24. [Crossref] [PubMed]
- Zhang P, Yang F, Luo Q, et al. miR-1284 Inhibits the Growth and Invasion of Breast Cancer Cells by Targeting ZIC2. Oncol Res 2019;27:253-60. [Crossref] [PubMed]
- Wang J, Ma W, Liu Y. Long non-coding RNA HULC promotes bladder cancer cells proliferation but inhibits apoptosis via regulation of ZIC2 and PI3K/AKT signaling pathway. Cancer Biomark 2017;20:425-34. [Crossref] [PubMed]
- Sabater L, Bataller L, Suarez-Calvet M, et al. ZIC antibodies in paraneoplastic cerebellar degeneration and small cell lung cancer. J Neuroimmunol 2008;201-202:163-5. [Crossref] [PubMed]
- Lu SX, Zhang CZ, Luo RZ, et al. Zic2 promotes tumor growth and metastasis via PAK4 in hepatocellular carcinoma. Cancer Lett 2017;402:71-80. [Crossref] [PubMed]
- Bawa PS, Ravi S, Paul S, et al. A novel molecular mechanism for a long non-coding RNA PCAT92 implicated in prostate cancer. Oncotarget 2018;9:32419-34. [Crossref] [PubMed]
- Chan DW, Liu VW, Leung LY, et al. Zic2 synergistically enhances Hedgehog signalling through nuclear retention of Gli1 in cervical cancer cells. J Pathol 2011;225:525-34. [Crossref] [PubMed]
- Gerstein M. Integrative database analysis in structural genomics. Nat Struct Biol 2000;7 Suppl:960-3. [Crossref] [PubMed]
- Troiano G, Guida A, Aquino G, et al. Integrative Histologic and Bioinformatics Analysis of BIRC5/Survivin Expression in Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences 2018;19:2664. [Crossref] [PubMed]
- Wang K, Ruan H, Song Z, et al. PLIN3 is up-regulated and correlates with poor prognosis in clear cell renal cell carcinoma. Urol Oncol 2018;36:343.e9-343.e19. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Cline MS, Craft B, Swatloski T, et al. Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci Rep 2013;3:2652. [Crossref] [PubMed]
- Güre AO, Stockert E, Scanlan MJ, et al. Serological identification of embryonic neural proteins as highly immunogenic tumor antigens in small cell lung cancer. Proc Natl Acad Sci U S A 2000;97:4198-203. [Crossref] [PubMed]
- Bidus MA, Risinger JI, Chandramouli GV, et al. Prediction of lymph node metastasis in patients with endometrioid endometrial cancer using expression microarray. Clin Cancer Res 2006;12:83-8. [Crossref] [PubMed]
- Fernebro J, Francis P, Eden P, et al. Gene expression profiles relate to SS18/SSX fusion type in synovial sarcoma. Int J Cancer 2006;118:1165-72. [Crossref] [PubMed]
- Shen ZH, Zhao KM, Du T. HOXA10 promotes nasopharyngeal carcinoma cell proliferation and invasion via inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci 2017;21:945-52. [PubMed]
- Inaguma S, Ito H, Riku M, et al. Addiction of pancreatic cancer cells to zinc-finger transcription factor ZIC2. Oncotarget 2015;6:28257-68. [Crossref] [PubMed]
- Pfister S, Schlaeger C, Mendrzyk F, et al. Array-based profiling of reference-independent methylation status (aPRIMES) identifies frequent promoter methylation and consecutive downregulation of ZIC2 in pediatric medulloblastoma. Nucleic Acids Res 2007;35:e51. [Crossref] [PubMed]
- Sakuma K, Kasamatsu A, Yamatoji M, et al. Expression status of Zic family member 2 as a prognostic marker for oral squamous cell carcinoma. J Cancer Res Clin Oncol 2010;136:553-9. [Crossref] [PubMed]
- Zhou J, Zhu G, Huang J, et al. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: A novel potential therapeutic target. Cancer Lett 2016;370:313-23. [Crossref] [PubMed]
- D'Amato C, Rosa R, Marciano R, et al. Inhibition of Hedgehog signalling by NVP-LDE225 (Erismodegib) interferes with growth and invasion of human renal cell carcinoma cells. Br J Cancer 2014;111:1168-79. [Crossref] [PubMed]
- Mercier S, Dubourg C, Garcelon N, et al. New findings for phenotype-genotype correlations in a large European series of holoprosencephaly cases. J Med Genet 2011;48:752-60. [Crossref] [PubMed]