Extent and characteristics of immune infiltration in clear cell renal cell carcinoma and the prognostic value
Introduction
It has been widely recognized that in addition to tumor cells, tumor tissue contains other important components such as vascular cells, stromal cells and immune cells, which all play a vital role in tumor development and progression. The tumor micro-environment is of great influence on tumor’s biological behavior. Therefore, the tumor tissue is more appropriate to be treated as an organ but not a mass of transformed epithelial cells (1-3). More and more researchers have been investigating into tumor micro-environment and gained more and more achievement revealing the effect of tumor micro-environment on various kinds of cancers (4,5).
Yoshihara et al. developed a new algorithm that can infer tumor cellularity using transcriptional profiles of cancer samples and it can estimate the relative amount of the stromal and immune cells that form the main non-tumor components by outputting ESTIMATE score as well as Stromal score and Immune score (6).
According to Yoshihara et al., clear cell renal cell carcinoma (ccRCC) has relatively high Stromal score and Immune score among common cancers (6). Kidney cancer contains several kinds of malignant diseases, and RCC is the most common form and accounts for 85% of Kidney cancers (7). Besides, ccRCC is the most common type of RCC. The result of Yoshihara et al.’s study indicates that the stromal and immune components may have important influence on ccRCC. To further confirm it and preliminary explore possible mechanisms we conducted this research on basis of The Cancer Genome Atlas (TCGA) and GEO data sets and revealed that Immune score is significantly associated with the stage, metastases, prognosis of RCC. What’s more, regulatory T cells (T-regs) may play a vital role in RCC progression and CXCL-1 and SAA1 may potentially take part in that process.
Methods
Search strategy
Gene expression profile as well as clinical data such as gender, age, stage, whether metastasis, survival and outcome for ccRCC patients was obtained from the TCGA data portal of KIRC (https://tcga-data.nci.nih.gov/tcga/) before March 30, 2019. Besides, all relevant datasets were identified by comprehensively searching NCBI Gene Expression Omnibus (GEO) datasets before March 30, 2019. The searching strategy was consisted of the following keywords: “Homo sapiens” and “Series” and “Expression profiling by array” and “renal cell carcinoma” and (“predict” or “prognosis” or “survival”). Inclusion and exclusion criteria: The inclusion criteria were as follows: (I) bio specimens were collected from patients with renal cell carcinoma; (II) containing at least 50 samples in each dataset; (III) including both clinical parameters [age, clinical stage (T) or grade] and overall survival (OS). The exclusion criteria were as follows: (I) duplicates of the previous eligible datasets; (II) datasets without sufficient and qualified data.
Estimate of immune infiltration in ccRCC
Immune scores and stromal scores were calculated by applying the ESTIMATE algorithm to the downloaded database (6). The CIBERSORT method, a computational approach to inferring leukocyte representation in bulk tumor transcriptomes (8).
Analysis of differentially expressed genes (DEGs)
Data analysis was performed using package limma (9). Fold change >1.5 and adj. P<0.05 was set as the cutoffs to screen for DEGs. Heatmaps and clustering were generated using an open source web tool ClustVis (10).
The protein-protein interaction (PPI) network was retrieved from STRING database (11) and reconstructed via Cytoscape software (12). Only individual networks of 10 or more nodes were included for further analysis. Networks of fewer than 10 nodes were excluded. The connectivity degree of each node of the network was calculated. Molecular COmplex DEtection (MCODE) was then used to find clusters based on topology to locate densely connected regions.
Statistics
Kaplan-Meier plots were generated to illustrate the relationship between patients’ overall survival and gene expression levels of DEGs. The relationship was tested by log-rank test. Enrichment analysis of DEGs Functional enrichment analysis of DEGs was performed by DAVID (The Database for Annotation, Visualization and Integrated Discovery) (13) to identify GO categories by their biological processes (BP), molecular functions (MF), or cellular components (CC). The DAVID database was also used to perform pathway enrichment analysis with reference from KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. False discovery rate (FDR) <0.05 was used as the cut-off. Correlation between Immune/Stromal scores, property of each kind of inferring leukocyte as well as each DEGs and stage, metastases and Fuhrman grade was conducted by Wilcox Test. Correlation was verified by Pearson correlation analysis. The scatter plot was made using GraphPad Prism7 software.
Results
Enrolled cases in TCGA and GEO
Gene expression profiles and clinical information (survival, stage and metastases) of all 537 (346 males and 191 females, 360 alive and 177 dead) ccRCC patients with initial pathologic diagnosis from the TCGA database was downloaded while gene expression profiles and clinical information (Fuhrman grade) of all 101 (59 males and 42 females) from GEO (GSE40435) was downloaded. The whole process of analysis was shown in the flow diagram (Figure 1).
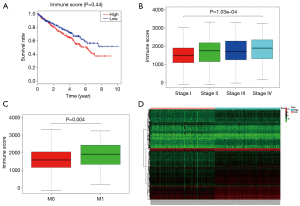
Immune scores but not stromal scores are significantly associated with OS, stage, metastases of ccRCC
The Immune and Stromal scores of each patient from TCGA database were obtained by applying ESTIMATE algorithm and all of the patients were divided into two groups based on the median of Immune/Stromal score. The results of Kaplan-Meier survival analysis showed that high Immune scores but not stromal scores are significantly associated with OS, stage, metastases of RCC (Figure 2A,B,C). The 537 RCC cases in TCGA database were divided into high/low groups based on the median of Immune score. A total of 512 genes were upregulated and 147 genes were downregulated in the high score than the low score group (fold change >1.5, P<0.05), as DEGs. Distinct gene expression profiles of cases belong to high and low Immune score group were showed in heatmap (Figure 2D).
Correlation of expression of individual DEGs in overall survival
To further explore the potential influence of individual DEGs on overall survival, we performed Kaplan-Meier survival analysis with data from TCGA database. Among the 659 DEGs upregulated/downregulated in the high-immune scores group, a total of 239 DEGs were shown to significantly predict disadvantage/advantage overall survival in log-rank test (P<0.05, selected genes are shown in Figure 3), into which we would further investigate.
Correlation of expression of individual survival significantly related genes in stage and metastases
We further investigated whether each of the 239 survival-significantly-related genes was significantly associated with the stage and metastases of ccRCC by applying Wilcox Test and found that 92 genes were significantly associated with stage of ccRCC and 198 with metastases (P<0.05, selected genes are shown in Figure 4).
Functional enrichment analysis of the 239 survival-significantly-related DEGs
We further performed Functional enrichment analysis of the 239 survival-significantly-related genes. Top gene ontology (GO) terms identified included T cell activation, T-regs activation, external side of plasma membrane and receptor ligand activity (Figure 5A). Besides, all pathways yielded from the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were related to immune response and the top one was cytokine-cytokine receptor interaction (Figure 5B).
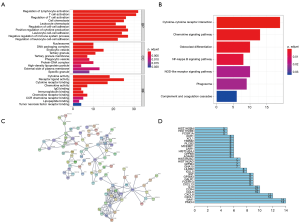
PPIs among genes of prognostic value
PPI network among 239 survival-significantly-related DEGs were obtained using the STRING tool respectively (Figure 5C). We recorded the genes with most connected nodes in the network (Figure 5D). As is shown, PMCH, SAA1 and several genes in CXCL-family turned up in top position, which means they played central role in both networks.
The central genes associated with survival, stage and metastases of ccRCC
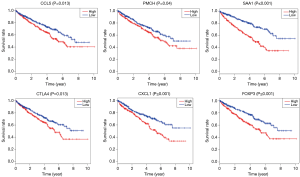
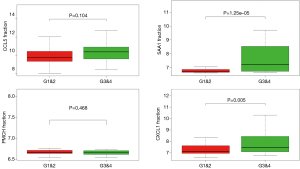
In order to identify the potential centralist genes in the development of ccRCC, we filtered the genes that had at least 10 connected nodes in PPI networks among 239 survival-significantly-related genes and were significantly associated with all three of survival, stage, metastases of ccRCC. Four genes remained meeting all the criterion: PMCH, SAA1, CXCL1, CCL5. Then, we verified our result in GEO database by assessing whether the four genes were associated with Fuhrman grade of ccRCC and revealed that CXCL1 and SAA1 were different in cases with low (I and II) grade and high (III and IV) grade. CCL5 and PMCH, however, was not (Figure 6).
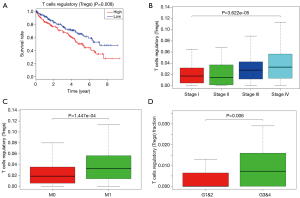
Proportion of T-regs is significantly negatively correlated to OS, stage and metastases of ccRCC
To explored the potential kind of immune cell that played the role in promoting ccRCC’s progression, we applied CIBERSORT method and revealed that the proportion of T-regs is significantly negatively correlated to OS (with the lowest P value), stage and metastases of ccRCC, which was in accordance with the result of FOXP3, a specific marker of T-reg (Figure 7). What’s more, proportion of T-regs was also different in cases with low (I and II) grade and high (III and IV) grade after being verified in GEO.
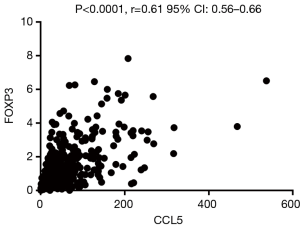
Foxp3 is significantly positively correlated with CCL5 but not CXCL1
CXCL-1 and CCL5 are both chemokine, which may play a vital role in recruiting T-regs. To further explore the possible relationship between the two chemokines and T-regs, related analysis were conducted between the expression of FOXP3 and CXCL1/CCL5 based on the RCC cases on TCGA database. It was revealed that FOXP3 is significantly positively correlated with CCL5 (Figure 8) but not CXCL1.
Discussion
Solid tumor is not only a mass of malignant tumor cells, but composed of stromal and immune cells as well. Next-generation sequencing has provided us with an unprecedented ability to study tumor (14). TCGA database has also been providing a lot of precious information for researchers and many valuable results have been generated based on it (15,16).
In this study, we assessed the association between immune infiltration and the clinical features as well as prognosis of ccRCC and the extent and characteristics of immune infiltration in ccRCC were also examined. The analysis was performed by applying ESTIMATE algorithm and CIBERSORT method based on the gene expression profile on TCGA database and validated using the data on GEO.
We revealed that the immune infiltration was significantly associated with higher stage, more chances of metastases and the poor prognosis of ccRCC. And we obtained some important immune-related DEGs that could predict the survival of ccRCC patients. The GO and KEGG analysis further revealed that the DEGs were involved in the processes of T cell activation, T-regs activation, external side of plasma membrane, receptor ligand activity and cytokine-cytokine receptor interaction.
Then we investigated which kind of immune cell play a vital role in the process of ccRCC progression and T-regs turned up in the analysis using CIBERSORT method. We confirmed that T-regs’ infiltration was also significantly associated with higher stage, more chances of metastases and the poor prognosis of ccRCC as well as higher Fuhrman grade.
We also found 4 central genes that had most connected nodes in PPI network and furthermore significantly associated with higher stage, more chances of metastases and the poor prognosis of ccRCC: PMCH, SAA1, CXCL1, CCL5, of which, CXCL-1 and SAA1 were significantly associated with Fuhrman grade of ccRCC according to data in GEO. To explored the potential relationship between the four central genes and T-regs, we searched relative literature and found it reported that CXCL1 and CCL5 could recruit T-regs in non-small cell lung cancer and pancreatic ductal adenocarcinoma respectively (17,18). Serum amyloid A (SAA) was identified as a mitogen for T-regs by a monocyte-dependent process mediated by IL-1β and IL-6 (19). What’s more, SAA1 was previews reported to have predictive value in prognosis of RCC (20).
Cause both methods are indirect measures of immune infiltration rather than direct detection, in order to minimize the bias of the algorithm, we use two methods at the same time so that we can find out the cross point between them and verify it again in the TCGA database and the GEO database to avoid the impact of the limitations and particularity of a single algorithm and a single database on the results.
T-regs play a vital role in tumor immunosuppression (21,22). Many studies have revealed its role in various cancers (23-26). Miska et al. (27) revealed that HIF-1α is a metabolic switch between glycolytic-driven migration and oxidative phosphorylation-driven immunosuppression of T-regs in glioblastoma. It is worth attention that HIF-1α also takes part in the development of ccRCC (28), which may be a potential mechanism by which T-regs influence ccRCC.
Chemokines are a family of small inflammatory cytokines that are able to induce chemotaxis in responsive cells (29). In tumor, chemokines as well as their receptors are of great importance in regulating cell trafficking in and out of the tumor microenvironment (TME) (30). CXCL-1 and CCL5 are both members of Chemokines families and have been reported to promote tumor progression in kinds of cancers (31,32). But there still few studies focusing on the relationship between them and ccRCC. According to our results, Lv et al.’s and Wang et al.’s studies, they may take part in the interaction between T-regs and ccRCC, especially CCL5 cause it is significantly positively correlated with foxp3, the marker of T-reg. what’s important the conjecture need to be confirmed by further basic research especially mechanism research.
What is an important strategy to lift the immunosuppression and achieve immune normalization is remodeling tumor immune microenvironment (TIME) and there has already arisen some reports (33). Since PD-1/PD-L1 blockade therapy Nivolumab has been approved (34), immunity therapy shows great potential in treatment of ccRCC. Our results provided some potential targets of immunity therapy, which are worth further research and may promote ccRCC treatment in the future.
Conclusions
Immune infiltration in RCC, especially the property of T-regs has a negative influence on ccRCC. We identified several immune relative proteins that may take part in the process—CXCL-1, SAA1, CCL5 and PMCH, among which, CCL5 may be the central proteins that mediate the T-regs’ participation in ccRCC progression.
Acknowledgments
Funding: The study was supported by the National Natural Science Foundation of China (Grants Nos. 81270685, 81771640 and 81672532) and Project of Nanjing Science and Technology Committee (201605001).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013;501:346-54. [Crossref] [PubMed]
- Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer 2019;19:65-81. [Crossref] [PubMed]
- Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer 2018;18:407-18. [Crossref] [PubMed]
- Kundu B, Bastos AR, Brancato V, et al. Mechanical Property of Hydrogel and the Presence of Adipose Stem Cells in Tumor Stroma Affect Spheroid Formation in 3D Osteosarcoma Model. ACS Appl Mater Interfaces 2019;11:14548-59. [Crossref] [PubMed]
- Tokunaga R, Naseem M, Lo JH, et al. B cell and B cell-related pathways for novel cancer treatments. Cancer Treat Rev 2019;73:10-9. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA -sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Metsalu T, Vilo J.. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 2015;43:W566-70. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Turajlic S, Sottoriva A, Graham T, et al. Resolving genetic heterogeneity in cancer. Nat Rev Genet 2019;20:404-16. [Crossref] [PubMed]
- Kumada K, Yasuda J. Evolutionary acquired robustness and vulnerability in cancer genome: negligible negative selection in carcinogenesis. Transl Cancer Res 2018;7:S445-8. [Crossref]
- Kraya AA, Maxwell KN, Wubbenhorst B, et al. Genomic signatures predict the immunogenicity of BRCA-deficient breast cancer. Clin Cancer Res 2019;25:4363-74. [Crossref] [PubMed]
- Lv M, Xu Y, Tang R, et al. miR141-CXCL1-CXCR2 signaling-induced Treg recruitment regulates metastases and survival of non-small cell lung cancer. Mol Cancer Ther 2014;13:3152-62. [Crossref] [PubMed]
- Wang X, Lang M, Zhao T, et al. Cancer-FOXP3 directly activated CCL5 to recruit FOXP3+Treg cells in pancreatic ductal adenocarcinoma. Oncogene 2017;36:3048-58. [Crossref] [PubMed]
- Nguyen KD, Macaubas C, Truong P, et al. Serum amyloid A induces mitogenic signals in regulatory T cells via monocyte activation. Mol Immunol 2014;59:172-9. [Crossref] [PubMed]
- Paret C, Schön Z, Szponar A, et al. Inflammatory protein serum amyloid A1 marks a subset of conventional renal cell carcinomas with fatal outcome. Eur Urol 2010;57:859-66. [Crossref] [PubMed]
- Wing JB, Tanaka A, Sakaguchi S. Human FOXP3+ Regulatory T Cell Heterogeneity and Function in Autoimmunity and Cancer. Immunity 2019;50:302-16. [Crossref] [PubMed]
- Togashi Y, Shitara K, Nishikawa H.. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat Rev Clin Oncol 2019;16:356-71. [Crossref] [PubMed]
- Lim CJ, Lee YH, Pan L, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut 2019;68:916-27. [Crossref] [PubMed]
- Chew V, Lai L, Pan L, et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc Natl Acad Sci U S A 2017;114:E5900-9. [Crossref] [PubMed]
- Matoba T, Imai M, Ohkura N, et al. Regulatory T cells expressing abundant CTLA-4 on the cell surface with a proliferative gene profile are key features of human head and neck cancer. Int J Cancer 2019;144:2811-22. [Crossref] [PubMed]
- Toker A, Nguyen LT, Stone SC, et al. Regulatory T Cells in Ovarian Cancer Are Characterized by a Highly Activated Phenotype Distinct from that in Melanoma. Clin Cancer Res 2018;24:5685-96. [Crossref] [PubMed]
- Miska J, Lee-Chang C, Rashidi A, et al. HIF-1α Is a Metabolic Switch between Glycolytic-Driven Migration and Oxidative Phosphorylation-Driven Immunosuppression of Tregs in Glioblastoma. Cell Rep 2019;27:226-37.e4. [Crossref] [PubMed]
- Schödel J, Grampp S, Maher ER, et al. Hypoxia, Hypoxia-inducible Transcription Factors, and Renal Cancer. Eur Urol 2016;69:646-57. [Crossref] [PubMed]
- Bian X, Xiao YT, Wu T, et al. Microvesicles and chemokines in tumor microenvironment: mediators of intercellular communications in tumor progression. Mol Cancer 2019;18:50. [Crossref] [PubMed]
- Weitzenfeld P, Ben-Baruch A. The chemokine system, and its CCR5 and CXCR4 receptors, as potential targets for personalized therapy in cancer. Cancer Lett 2014;352:36-53. [Crossref] [PubMed]
- Wei LY, Lee JJ, Yeh CY, et al. Reciprocal activation of cancer-associated fibroblasts and oral squamous carcinoma cells through CXCL1. Oral Oncol 2019;88:115-23. [Crossref] [PubMed]
- Li L, Xu L, Yan J, et al. CXCR2-CXCL1 axis is correlated with neutrophil infiltration and predicts a poor prognosis in hepatocellular carcinoma. J Exp Clin Cancer Res 2015;34:129. [Crossref] [PubMed]
- Wang H, Tang Y, Fang Y, et al. Reprogramming Tumor Immune Microenvironment (TIME) and Metabolism via Biomimetic Targeting Codelivery of Shikonin/JQ1. Nano Lett 2019;19:2935-44. [Crossref] [PubMed]
- Opdivo (nivolumab) injection, for intravenous use. Princeton, NJ: Bristol-Myers Squibb Company, 2018. Available online: (accessed Jan 7, 2019).https://packageinserts.bms. com/pi/pi_opdivo.pdf