
Prostate cancer biology & genomics
The burden of prostate cancer
Prostate cancer is the most common cancer for males in the UK. About 1 in 6 men are predicted to develop prostate cancer over their lifetime (1). In 2016, 11,631 patients died from prostate cancer in the UK (2). Until recently, the causes of prostate cancer were unclear owing to significant heterogeneity in samples between patients, and even within the same patient. However, the development of gene sequencing technology in the past few decades has revolutionized the study of genomics.
The origin of prostate cancer
The prostate is a walnut sized glandular structure surrounded by a collagenous capsule, Denonviller’s fascia and neurovascular bundle. Within the structure lies the urethra in which glandular fluid drains from the various areas of the prostate. Most of the glandular elements of the prostate lie within the peripheral zone, which is held together in a fibrous prostatic capsule within a stromal and collagen meshwork. It is known from radical prostatectomy samples that 68% prostate cancer arise from the peripheral zone (3-5).
Within the epithelial layer lies three cell types: the luminal acinar cells which secrete glandular fluid involved in semen production, basal cells which line the basal layer and are integral to survival of the luminal cells, and finally sparsely populated neuroendocrine cells which are involved in paracrine and endocrine signalling. Experimental difficulties isolating the underlying cell type responsible for the transformative change in prostate cancer means the origin still eludes researchers. The loss of the basal layer forms part of the diagnostic criteria for prostate cancer. Some studies have also suggested that a separate type of cell termed “intermediate” cells may be involved in carcinogenesis (6). However, no study has been able to prove beyond doubt the origin of prostate cancer and it is possible, given the significant inter- and intra-tumoural heterogeneity, that prostate cancer can arise from multiple cell types.
Genome-wide association studies and single nucleotide polymorphisms (SNPs)
Prostate cancer risk factors are very varied. Age, ethnicity and family history are the most well recognized risk factors for prostate cancer (7-9), although environment, lifestyle and sexual history have also been implicated (10-13). Since the completion of the Human Genome Project (14,15) and advent of genome wide association studies, many studies have attempted to identify susceptible loci for prostate cancer. Initial family linkage and twin studies failed to find reproducible results owing to the heterogeneity of prostate cancer tumorigenesis. Genes such as HOXB13 (16), BRCA1 (17), BRCA2 (18,19) were identified to confer significantly increased risk in prostate cancer development, aggression and are inherited in an autosomal dominant fashion with high penetrance, but this only accounted for a small percentage of familial prostate cancer. A recent review by Rubin and Demichaelis in 2018 (20) described several common genomic alterations in prostate cancer in which PTEN, RB1, TP53, AR and C-MYC are the most frequently implicated.
Other studies have identified further susceptible genes and foci, but the risk did not always translate to disease development. In a meta-analysis and GWAS of 24,395 cases and 24,726 controls the SNP rs11672691 showed evidence of replication with genome wide significance (21). However, rs8khie87391, located the same region as rs11672691, was associated with prostate cancer, but not found to be significant in another GWAS of 2,393 cases and 1,222 controls (22). Common alterations across all prostate cancer samples are still elusive and reflect patient heterogeneity.
In 2018, Schumacher et al. (23) published a meta-analysis of 79,194 prostate cancer samples and 61,112 control samples which identified 63 new prostate cancer susceptibility loci. Around half of the data generated for this meta-analysis was generated by using OncoArray, a microarray designed to detect around 600,000 SNPs. This study builds upon previous GWAS results which identified approximately 100 prostate cancer susceptibility foci (24,25) and led to the development of the BARCODE 1 trial in 2017 predicting prostate cancer risk from a saliva test (26).
It is widely accepted that prostate cancer is pathologically multi-focal following autopsy studies by Djavan et al. (27) in 1999 showing 66% of cases were multifocal and 33% had a single focus of disease. Early genomic studies by Liu et al. (28) in 2009 using genome-wide SNP analysis of 94 anatomically separate cancer sites from 30 patients who passed away from metastatic prostate cancer found no relationship between the anatomic site of metastasis and the genomic copy number change pattern, suggesting a monoclonal origin of metastatic prostate cancer. However, subsequent genomic studies suggest that multifocal prostate cancer is a result of branching evolution, subsequent differentiation and ultimately mixing clonal populations. This gives evidence to the theory that cancer exhibits a field effect whereby surrounding morphologically normal tissue also exhibits high levels of mutations consistent with cancer cells despite being morphologically normal (29-32). It is possible that some patients may have monoclonal expansions whilst others have mixed metastatic clones. Further work is required in this area to understand the progression of metastatic prostate cancer.
Theories of treating the index lesion to reduce cancer burden has led to focal therapies gaining traction within parts of the urology community (33). Prospective studies (34,35) and meta-analysis (36) of morbidity data in whole gland versus focal therapy treatments have shown significantly reduced complication rates, although comparative toxic end points are still lacking (37). At present, UK National Guidelines continue to recommend whole gland therapy for intermediate to high risk patients (38).
Proteomic and genomic diagnostic and prognostic factors
Prostate cancer is often inherently slow-growing, and diagnosis is complicated by the presence of clinically insignificant cancer. Prostate specific antigen (PSA) was first purified in 1979 by Wang et al. (39). The discovery of PSA and subsequent studies as a serum biomarker created a new era of testing in prostate cancer. Unfortunately, PSA can be raised or under-represented in many benign conditions unrelated to prostate cancer (Table 1).

Full table
Due to the inaccuracies and ethical issues of overtreating patients with clinically insignificant prostate cancer (49), studies have attempted to validate the use of PSA in general screening. In 2013, Vickers et al. (50) performed retrospective PSA testing on archival samples from the Malmo Preventive Project in Sweden from the 1970s and compared them to previous PSA results. They concluded that measurements of PSA concentration from early to mid-life can identify small groups of men at increased metastatic risk several decades later. Their results were widely reported in the media as evidence to offer screening for men in the 40s (51). However, the study results and interpretation were mired in controversy including questions over the stability of archival samples, changes in the PSA assay over time and the suitability of taking multiple PSA tests (the authors recommended at least 3) over their lifetime to screen for a small number of men. What is interesting is that an article published by Wilt and Ahmed (52) on prostate cancer screening in the same edition of BMJ recommended that “By choosing not to have the PSA test you can live a similar length of life, have little to no difference in your risk of dying from prostate cancer, and avoid the harms associated with tests, procedures, and treatments”. In 2018, Ilic et al. (53) performed a systematic review and meta-analysis of five randomized control trials involving a total of 721,718 men comparing PSA screening with routine care. He concluded that screening may lead to a small reduction in disease-specific mortality over 10 years but has no effect on overall mortality.
At present, PSA is used both as part of a diagnostic algorithm used for at risk patients (54) as well as for assessing biochemical recurrence but, due to its poor specificity, alternative clinical scoring systems including Gleason score, TMN staging, CAPRA score (55), imaging and histopathological examination should be incorporated when assessing the holistic long-term risk for patients and deciding treatment as a multidisciplinary team.
With improvements in proteomics and genomics, multiple diagnostic and prognostic indicators are now available commercially to assist in clinical decision making (Table 2).
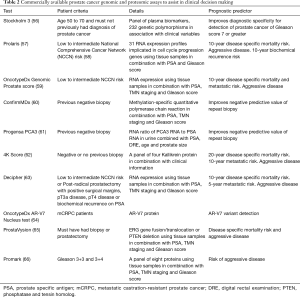
Full table
It is important to note that, at present, the gold standard is still biopsy and histopathology examination, but many of these tests will provide further indications whether patients should receive invasive testing or treatment.
Androgen receptor (AR): the molecular basis of CRPC treatments
The relationship between androgens and prostate cancer was first discovered in a landmark study by Huggins and Hodges (67) in 1941, and they were subsequently awarded the Nobel Prize in Medicine and Physiology 1966 (68). Further work revealed that this is largely driven by the AR. In recurrent prostate cancer, androgen deprivation therapy (ADT) is utilized with good results. However, ultimately, most cases will progress to castration-resistant prostate cancer (CRPC) due to extragonadal sources of androgens, AR overexpression and amplification (69), mutation and variants (70) or ligand-independent transactivation. A small number of patients will develop AR independent metastatic castration-resistant prostate cancer (mCRPC) which is linked to more aggressive phenotypes.
AR structure and signalling
AR is a ligand-activated transcription factor found on the X chromosome and dysregulation is associated with prostate cancer. The wild-type AR contains an N-terminus, DNA binding domain, hinge region and ligand-binding domain.
AR ligand binding domain
The ligand-binding domain is involved in the activation of AR. Unliganded AR resides within the cytoplasm in combination with heat shock proteins. Androgens enter the cell via diffusion and bind to the C-terminal ligand-binding domain of the receptor, causing a conformational change and dissociation of the heat shock protein complex, revealing a nuclear localization sequence (NLS). The AR with an exposed NLS localizes to the nucleus and binds to a variety of androgen response elements throughout the genome, resulting in modulation of gene expression. Modern CRPC treatment utilizes multiple modalities as the range of patients varies significantly from those with asymptomatic persistently high PSA to those with metastatic lesions (71). Current treatments targeting the AR receptor aim to disrupt this signalling pathway at various points.
AR N-terminus
In 1995, Jenster et al. (72) performed a series of AR N-terminal deletions which identified that amino acid residues 1–485 (Transcription Activating Unit 1) were required for full AR activity via the ligand-activating pathway, whilst preservation of amino acids residues 360–528 (Transcription Activating Unit 5) were sufficient to allow transcription by the AR even when the ligand-binding domain was deleted. The N-terminus also contains a CAG and GGC trinucleotide repeat polymorphism, which varies in length (73). The significance of variations in AR CAG polymorphism remains controversial, with some studies associating shorter repeat lengths with higher incidence of prostate cancer in African American men versus Caucasian men (74-77), but others not observing the same results in European populations (78,79). In 1994, Chamberlain et al. (80) found that increasing CAG repeat length reduced transactivation of transcription factors but did not eliminate AR activity.
AR DNA binding domain
The DNA binding domain consists of two zinc molecules surrounded by nine cysteine molecules. The two “finger-like” zinc projections function to provide specificity to DNA binding as well as a dimerization interface to stabilize binding (81,82).
AR hinge region
The hinge region is involved in the regulation of AR activity. It has a role in differentiation between classical and selective androgen response elements and in post-translational modifications (83).
AR mutations
AR mutations are rare in the early stages of prostate cancer but increases in mCRPC. In 2015, Robinson et al. (84) published matched genome wide sequencing, germline and transcriptomics data of 150 mCRPC samples which identified 62.7% of all mCRPC samples harboured mutations in AR. Interestingly, their data showed a relative minority of mutations were within exon 1 (N-terminal and CAG polymorphism) and showed that most mutations focused in exon 4–8 (ligand-binding domain). The My Cancer Genome database manually aggregates data from multiple papers and databases including the Catalogue of Somatic Mutations in Cancer (85) and shows that L702H, W742C, H875Y, F877L, T878A and AR-V7 are the most common AR mutations. Other open access resources such as cBioPortal also exists nowadays to aggregate cancer data (86,87).
Anti-androgen therapy
AR mutations have been implicated in bicalutamide failure. Bicalutamide is a competitive inhibitor of testosterone and dihydrotestosterone binding to the AR. In 2002, Steketee et al. (88) looked at ligand responsiveness to AR mutants using a MMTV-LUC reporter and found that bicalutamide did not activate wild-type AR or the AR mutants H874Y, T877A, T877C, T877G or T8775. While they did not show the data for the other 877 variants they tested, they state that bicalutamide did not activate any of the other 877 variants either. Interestingly, other studies have found that first generation anti-androgens such as bicalutamide, flutamide and nilutamide can have an agonist effect in some patients after several years of treatment, and conversely withdrawal of the drug appears to initially reduce tumour burden—a phenomenon coined as ‘androgen withdrawal syndrome’. This was experimentally shown when Tan et al. (89) demonstrated anti-androgen transactivation of AR using a luciferase reporter with AR mutants from mouse xenograph CWR22. They found that treatment with hydroxyflutamide (an early anti-androgen) at 10 nMol caused a 4- and 6-fold increase in transcriptional activity for H874Y and T877A mutants respectively compared to wild-type AR. However, total transcription activity was still only 15–20% of the maximal activity elicited by testosterone and dihydrotestosterone. They also noted that dehydroepiandrosterone (secreted by the adrenals) at 1 nMol, 10 nMol and 100 nMol stimulated an increase of 2-, 3- and 8-fold H874Y transcription activity compared to wild-type AR. In 2003, Hara et al. (90) cultured androgen-dependent LNCaP-FGC human cells with bicalutamide. They found that after 6–13 weeks, bicalutamide treatment increased PSA levels and growth in these LNCaPs. Subsequent sequencing of AR showed new mutations in W741C and W741L in the ligand-binding domain. Interestingly, hydroxyflutamide inhibited growth in these mutated LNCaP cells. This suggests that prolonged therapy with bicalutamide can cause AR mutations which cause bicalutamide to have agonist effects.
Enzalutamide is an anti-androgen capable of exerting action on three different stages of AR signalling. Firstly, it is a competitive AR inhibitor which binds to the ligand binding domain without triggering AR downstream signalling, thereby stopping androgens activating AR. It also has inhibitory actions on AR nuclear translocation and AR DNA binding within the nucleus (91). In 2013, Korpal et al. (92) and Joseph et al. (93) each generated different resistant LNCaP cell lines to overexpress AR and discovered a mutation at F876L within the AR ligand-binding domain which contributes to resistance to enzalutamide and apalutamide. Additionally, Korpal et al. observed that LNCaPs containing F876L were able to bypass the enzalutamide inhibition of AR nuclear translocation.
CYP17 inhibitor
Abiraterone irreversibly and selectively blocks cytochrome P450 17A1 (steroid 17α-monooxygenase) throughout the body, including those located within the adrenal cortex and the gonadal tissues. It also acts upon prostate cancer tissue itself and stops endogenous androgen production. As this drug is unselective for certain tissue groups, it effectively disrupts the hypothalamic-pituitary-adrenal axis, causing an increase in adrenocorticotrophic hormone and reduction in serum cortisol. Concurrent prednisolone treatment is therefore recommended for all patients using abiraterone (94,95).
Immunotherapy
The landmark discovery of dendritic cells in 1973 by Steinman et al. (96) led to the development of Sipuleucel-T immunotherapy and led to the first posthumous award in 2011 of the Nobel Prize in Medicine and Physiology for 50 years (97).
Sipuleucel-T immunotherapy is a therapeutic cancer vaccine designed to exploit the inherent nature of dendritic cells and their antigen presenting abilities. Antigen presenting cells are harvested from the patient, centrifuged and cultured with a combination of prostatic acid phosphatase and granulocyte-macrophage colony-stimulating factor antigen. The activated cells are infused back into the patient and recruit T-cells against prostate cancer cells (98). The exact mechanism of action once the cells are returned to the patient remains unknown. The IMPACT trial of 512 asymptomatic or minimally symptomatic men with mCRPC has shown that Sipuleucel-T has a statistically significant median improvement in survival of 4.1 months compared to control arms (99). However, each added month of survival is estimated to be at an average cost of $22,683 USD (100).
Chemotherapy
Failure of ADT or immunotherapy typically meant treatment with systemic chemotherapy. Hitchings and Elliot first developed rational molecules which lead to chemotherapy and they were awarded the Nobel Prize in Medicine and Physiology 1988 (101).
Docetaxel is one of the most commonly used chemotherapy drugs in prostate cancer. Mechanistically it is thought to inhibit cell division by acting on the tubulin network. Docetaxel also has cytoplasmic and nuclear activity against the AR, although the mechanism is not well understood (102). Large randomized control trials such as the CHAARTED and STAMPEDE trials have shown statistically significant improvements in median survival of 13.6 months (103) and 10 months respectively. The STAMPEDE trial further evaluated combinations of docetaxel and zoledronic acid together versus standard of care alone and concluded that zoledronic acid showed no evidence of survival improvement, whilst docetaxel did, albeit associated with an increase in adverse events (104).
Several new molecular therapeutics are currently under clinical trials including cycline-dependent kinase 4 and 6 (CDK4/6) inhibitors palbociclib, ribociclib, abemaciclib, poly adenosine diphosphate ribose polymerase (PARP) inhibitors olaparib, veliparib and talazoparib, and phosphoinositide 3-kinase (PI3K) inhibitors buparlisib and alpelisib.
In 2017, Goodall et al. (105) reported results from a prospective trial of serial circulating cell free-DNA of patients being treated with olaparib in a phase II clinical trial. By monitoring the change in gene expression and sequencing, they were able to demonstrate the emergence of mutations at progression that likely resulted in drug resistance compared to pre-treatment samples. This can have implications in predicting which patients may respond to certain drug treatments. It is hoped that in the future, biomarkers which allow clinicians to predict the effects of drug treatments in individuals will be available.
The treatments described here are not exhaustive and many are currently still in clinical trials. However, the mainstay of current CRPC treatment remains centred around targeting the AR.
Convergence towards personalized precision medicine
It is evident that a holistic picture of cancer is required given the significant heterogeneity between patients. Difficulties remain identifying the cell line responsible for development of prostate cancer. Whilst localized prostate cancer may develop from luminal acinar cells, it is possible there is an underlying stem cell population (such as intermediate cells) which may develop into CRPC and may explain drug resistance and treatment failure. GWAS studies have shown significant inter-tumoural, intra-tumoural and inter-patient heterogeneity. However, most mutations centre around AR, ETS Family, TP53, PTEN, C-MYC and FOXA1. Some studies have suggested that mCRPC may originate from a single clonal population. Genomic and proteomic studies have led to improvements in tests for prognostic factors, but these tests are only useful for certain situations and cannot predict every patient’s course. Often tests give rise to a probability of a risk of relapse or aggressive disease but these outcomes do not always occur. PSA on its own remains a poor diagnostic and prognostic marker.
Several therapeutics targeting AR are available, but due to AR overexpression, variants, mutations and other androgen production pathways, patients who relapse inevitably progress to CRPC despite AR inhibition. Not all patients react the same way to therapeutics and some patients are more sensitive to certain therapeutics than others. As cancer research increasingly embraces multi-disciplinary teams these bring together expertise in genomics, proteomics, radiomics, bioinformatics, clinical knowledge and surgery to translate medicine from the laboratory to the holistic patient.
Acknowledgments
We would like to thank our colleagues from University College London and Imperial College Healthcare NHS Trust for bringing together the manuscripts for this special edition.
Funding: This research was supported by National Institutes of Health Research Biomedical Research Centre Imperial College London, Cancer Research UK, the Imperial Prostate team headed by Professor Hashim U. Ahmed and University College London.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Martin J. Connor, Saiful Miah, Taimur T. Shah, Hashim U. Ahmed) for the series “Prostate Imaging and Focal Therapy” published in Translational Andrology and Urology. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: The series “Prostate Imaging and Focal Therapy” was commissioned by the editorial office without any funding or sponsorship. MJC served as the unpaid Guest Editor of the series. MJC is funded by the Wellcome Trust. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Smittenaar CR, Peterson KA, Steward K, et al. Cancer incidence and mortality projections in the UK until 2035. Br J Cancer 2016;115:1147-55. [Crossref] [PubMed]
- Cancer Research UK, Prostate cancer statistics, 2016 [online]. Available online: [accessed 17th May 2019].https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/prostate-cancer
- McNeal JE. Regional morphology and pathology of the prostate Am J Clin Pathol 1968;49:347-57. [Crossref] [PubMed]
- McNeal JE. Normal histology of the prostate. Am J Surg Pathol 1988;12:619-33. [Crossref] [PubMed]
- McNeal JE, Redwine EA, Freiha FS, et al. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol 1988;12:897-906. [Crossref] [PubMed]
- Xin L. Cells of origin for cancer: an updated view from prostate cancer. Oncogene 2013;32:3655-663. [Crossref] [PubMed]
- Bell KJ, Del Mar C, Wright G, et al. Prevalence of incidental prostate cancer: A systematic review of autopsy studies. Int J Cancer 2015;137:1749-57. [Crossref] [PubMed]
- Bratt O, Drevin L, Akre O, et al. Family History and Probability of Prostate Cancer, Differentiated by Risk Category: A Nationwide Population-Based Study J Natl Cancer Inst 2016;108:djw110. [Crossref] [PubMed]
- Kheirandish P, Chinegwundoh F. Ethnic differences in prostate cancer Br J Cancer 2011;105:481-5. [Crossref] [PubMed]
- Ferrís-I-Tortajada J, Berbel-Tornero O, Garcia-i-Castell J, et al. Non-dietary environmental risk factors in prostate cancer. Actas Urol Esp 2011;35:289-95. [PubMed]
- Downer MK, Batista JL, Mucci LA, et al. Dairy intake in relation to prostate cancer survival. Int J Cancer 2017;140:2060-9. [Crossref] [PubMed]
- Li H, Stampher MJ, Hollis JB, et al. A Prospective Study of Plasma Vitamin D Metabolites, Vitamin D Receptor Polymorphisms and Prostate Cancer. PLoS Med 2007;4:e103. [Crossref] [PubMed]
- Jian Z, Ye D, Chen Y, et al. Sexual Activity and Risk of Prostate Cancer: A Dose-Response Meta-Analysis. J Sex Med 2018;15:1300-9. [Crossref] [PubMed]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921. [Crossref] [PubMed]
- Venter JC, Adams MD, Myers EW, et al. The Sequence of the Human Genome Science 2001;291:1304-51. [Crossref] [PubMed]
- Xu J, Lange EM, Zheng SL, et al. HOXB13 is a susceptibility gene for prostate cancer: results from the International Consortium of Prostate Cancer Genetics (ICPCG). Hum Genet 2013;132:5-14. [Crossref] [PubMed]
- Mersch J, Jackson MA, Park M, et al. Cancers Associated with BRCA1 and BRCA2 Mutations other than Breast and Ovarian. Cancer 2015;121:269-75. [Crossref] [PubMed]
- Taylor RA, Fraser M, Livingstone J, et al. Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat Commun 2017;8:13671. [Crossref] [PubMed]
- Castro E, Eeles RA. The role of BRCA1 and BRCA2 in prostate cancer Asian J Androl 2012;14:409-14. [Crossref] [PubMed]
- Rubin MA, Demichaelis F. The Genomics of Prostate Cancer: emerging understanding of technological advances. Mod Pathol 2018;31:S1-11. [Crossref] [PubMed]
- Amin Al Olama A, Kote-Jarai Z, Schumacher FR, et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet 2013;22:408-15. [Crossref] [PubMed]
- Hsu FC, Sun J, Wiklund F, et al. A novel prostate cancer susceptibility locus at 19q13. Cancer Res 2009;69:2720-3. [Crossref] [PubMed]
- Schumacher FR, Olama AAA, Berndt S, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet 2018;50:928-36. [Crossref] [PubMed]
- Eeles RA, Olama AAA, Berndt S, et al. Prostate cancer meta-analysis from more than 145,000 men to identify 65 novel prostate cancer susceptibility loci. J Clin Oncol 2017;35:abstr 1.
- Eeles RA, Olama AAA, Beniloch S, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 2013;45:385-91. [Crossref] [PubMed]
- ClinicalTrials.gov, The BARCODE 1 Study (BARCODE1Pilot), US National Library of Medicine 2017 [online]. Available online: [accessed: 20th May 2019] [page last updated 27th February 2019].https://clinicaltrials.gov/ct2/show/NCT03158922
- Djavan B, Susani M, Bursa B, et al. Predictability and Significance of Multifocal Prostate Cancer in the Radical Prostatectomy Specimen. Tech Urol 1999;5:139-42. [Crossref] [PubMed]
- Liu W, Laitinen S, Khan S, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med 2009;15:559-65. [Crossref] [PubMed]
- Cooper CS, Eeles R, Wedge DC, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue Nat Genet 2015;47:367-72. [Crossref] [PubMed]
- Løvf M, Zhao S, Axcrona U, et al. Multifocal Primary Prostate Cancer Exhibits High Degree of Genomic Heterogeneity Eur Urol 2019;75:498-505. [Crossref] [PubMed]
- Wei L, Wang J, Lampert E, et al. Intratumoural and Intertumoural Genomic Heterogeneity of Multifocal Localized Prostate Cancer Impacts Molecular Classifications and Genomic Prognosticators. Eur Urol 2017;71:183-92. [Crossref] [PubMed]
- Su F, Zhang W, Zhang D, et al. Spatial Intratumour Genomic Heterogeneity within Localized Prostate Cancer Revealed by Single-nucleus Sequencing. Eur Urol 2018;74:551-9. [Crossref] [PubMed]
- Ahmed HU. The Index Lesion and the Origin of Prostate Cancer. N Engl J Med 2009;361:1704-6. [Crossref] [PubMed]
- Ahmed HU, Hindley RG, Dickinson L, et al. Focal therapy for localised unifocal and multifocal prostate cancer: a prospective development study. Lancet Oncol 2012;13:622-32. [Crossref] [PubMed]
- Barret E, Ahallal Y, Sanchez-Salas R, et al. Morbidity of Focal Therapy in the Treatment of Localized Prostate Cancer. Eur Urol 2013;63:618-22. [Crossref] [PubMed]
- Valerio M, Ahmed HU, Emberton M, et al. The Role of Focal Therapy in the Management of Localised Prostate Cancer: A Systematic Review. Eur Urol 2014;66:732-51. [Crossref] [PubMed]
- van der Poel HG, van der Bergh RCN, Briers E, et al. Focal Therapy in Primary Localised Prostate Cancer: The European Association of Urology Position in 2018. Eur Urol 2018;74:84-91. [Crossref] [PubMed]
- National Institute for Health and Care Excellence. Prostate cancer: diagnosis and management, 2014, [online]. Available online: [accessed 18th February 2019].https://www.nice.org.uk/guidance/cg175/chapter/1-Recommendations#assessment-2
- Wang MC, Valenzuela LA, Murphy GP, et al. Purification of a Human Prostate Specific Antigen. Invest Urol 1979;17:159-63. [PubMed]
- Oesterling JE, Jacobsen SJ, Chute CG, et al. Serum prostate-specific antigen in a community-based population of healthy men. Establishment of age-specific reference ranges. JAMA 1993;270:860-4. [Crossref] [PubMed]
- DeAntoni EP, Crawford ED, Oesterling JE, et al. Age- and race-specific reference ranges for prostate-specific antigen from a large community-based study. Urology 1996;48:234-9. [Crossref] [PubMed]
- Beebe-Dimmer JL, Faerber GJ, Morgenstern H, et al. Body composition and serum prostate-specific antigen: review and findings from Flint Men’s Health Study. Urology 2008;71:554-60. [Crossref] [PubMed]
- Adhyam M, Gupta AK. A Review on the Clinical Utility of PSA in Cancer Prostate. Indian J Surg Oncol 2012;3:120-9. [Crossref] [PubMed]
- Chybowski FM, Bergstrath EJ, Oesterling JE. The effect of digital rectal examination on the serum prostate specific antigen concentration: results of a randomized study. J Urol 1992;148:83-6. [Crossref] [PubMed]
- Tchetgen MB, Song JT, Strawderman M, et al. Ejaculation increases the serum prostate-specific antigen concentration. Urology 1996;47:511-6. [Crossref] [PubMed]
- Gamé X, Vincendeau S, Palascak R, et al. Total and Free Serum Prostate Specific Antigen Levels during the First Month of Acute Prostatitis. Eur Urol 2003;43:702-5. [Crossref] [PubMed]
- Oesterling JE, Rice DC, Glenski WJ, et al. Effect of cystoscopy, prostate biopsy, and transurethral resection of prostate on serum prostate-specific antigen concentration. Urology 1993;42:276-82. [Crossref] [PubMed]
- Aliasgari M, Soleimani M, Hosseini Moghaddam SM. The effect of acute urinary retention on serum prostate-specific antigen level. Urol J 2005;2:89-92. [PubMed]
- Loeb S, Bjurlin MA, Nicholson J, et al. Overdiagnosis and Overtreatment of Prostate Cancer. Eur Urol 2014;65:1046-55. [Crossref] [PubMed]
- Vickers AJ, Ulmert D, Sjoberg DD, et al. Strategy for detection of prostate cancer based on relation between prostate specific antigen at age 40-55 and long term risk of metastasis: case-control study. BMJ 2013;346:f2023. [Crossref] [PubMed]
- Roberts M. Prostate cancer: case to test men in their 40s, British Broadcasting Corporation 2013, [online]. Available online: [accessed 23rd May 2019].https://www.bbc.co.uk/news/health-22164449
- Wilt TJ, Ahmed HU. Prostate cancer screening and the management of clinically localised disease. BMJ 2013;346:f325. [Crossref] [PubMed]
- Ilic D, Djulbegovic M, Jung JH, et al. Prostate cancer screening with prostate-specific antigen (PSA) test: a systematic review and meta-analysis BMJ 2018;362:k3519. [Crossref] [PubMed]
- Tikkinen KAO, Dahm P, Lytvyn L, et al. Prostate cancer screening with prostate-specific antigen (PSA) test: a clinical practice guideline. BMJ 2018;362:k3581. [Crossref] [PubMed]
- Cooperberg MR, Pasta DJ, Elkin EP, et al. The UCSF Cancer of the Prostate Risk Assessment (CAPRA) Score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol 2005;173:1938-42. [Crossref] [PubMed]
- Ström P, Nordstrom T, Aly M, et al. The Stockholm-3 Model for Prostate Cancer Detection: Algorithm Update, Biomarker Contribution, and Reflex Test Potential. Eur Urol 2018;74:204-10. [Crossref] [PubMed]
- Cuzick J, Berney DM, Fisher G, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer 2012;106:1095-9. [Crossref] [PubMed]
- National Comprehensive Cancer Network, NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Prostate Cancer, Version 4, 2018 [online]. Available online: [last accessed 18th February 2019].https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- Van Den Eeden SK, Lu R, Zhang N, et al. A Biopsy-based 17-gene Genomic Prostate Score as a Predictor of Metastases and Prostate Cancer Death in Surgically Treated Men with Clinically Localised Disease. Eur Urol 2018;73:129-38. [Crossref] [PubMed]
- van Kessel KE, Beukers W, Lurkin I, et al. Validation of a DNA Methylation-Mutation Urine Assay to Select Patients with Hematuria for Cystoscopy. J Urol 2017;197:590-5. [Crossref] [PubMed]
- Durand X, Moutereau S, Xylinas E, et al. ProgensaTM PCA3 test for prostate cancer. Expert Rev Mol Diagn 2011;11:137-44. [Crossref] [PubMed]
- Zappala SM, Scardino PT, Okrongly D, et al. Clinical performance of the 4Kscore Test to predict high-grade prostate cancer at biopsy: A meta-analysis of us and European clinical validation study results. Rev Urol 2017;19:149-55. [PubMed]
- Magi-Galluzzi C, Yousefi K, Haddad Z, et al. Validation of the Decipher prostate cancer classifier for predicting 10-year preoperative metastasis from analysis of diagnostic needle biopsy specimens. J Clin Oncol 2017;34:abstr 59.
- Scher HI, Lu D, Schreiber NA, et al. Association of AR-V7 on Circulating Tumour Cells as a Treatment-Specific Biomarker with Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol 2016;2:1441-9. [Crossref] [PubMed]
- Blume-Jensen P, Berman DM, Rimm DL, et al. Development and Clinical Validation of an In Situ Biopsy-Based Multimarker Assay for Risk Stratification in Prostate Cancer. Clin Cancer Res 2015;21:2591-600. [Crossref] [PubMed]
- Bostwick Laboratories, Prostavysion, 2019, [online]. Available online: [last accessed 23rd May 2019].https://www.bostwicklaboratories.com/services/laboratory-services/urologic-pathology/ProstaVysion.aspx
- Huggins C, Hodges CV. The Effect of Castration, of Estrogen and of Androgen Injection on Serum Phosphatases in Metastatic Carcinoma of the Prostate. Cancer Res 1941;1:293-7.
- NobelPrize.org, Nobel Prize in Medicine and Physiology 1966, Nobel Media AB 2019 [online]. Available online: [accessed 25th January 2019].https://www.nobelprize.org/prizes/medicine/1966/summary/
- Linja MJ, Savinainen KJ, Saramaki OR, et al. Amplification and Overexpression of Androgen Receptor Gene in Hormone-Refractory Prostate Cancer. Cancer Res 2001;61:3550-5. [PubMed]
- Brooke GN, Bevan CL. The Role of Androgen Receptor Mutations in Prostate Cancer Progression. Curr Genomics 2009;10:18-25. [Crossref] [PubMed]
- Rathkopf D, Scher HI. Androgen Receptor Antagonists in Castration-Resistant Prostate Cancer. Cancer J 2013;19:43-9. [Crossref] [PubMed]
- Jenster G, van der Korput HA, Trapman J, et al. Identification of Two Transcription Activation Units in the N-terminal Domain of the Human Androgen Receptor. J Biol Chem 1995;270:7341-6. [Crossref] [PubMed]
- Nelson KA, White JS. Androgen Receptor CAG Repeats and Prostate Cancer Am J Epidemiol 2002;155:883-90. [Crossref] [PubMed]
- Irvine RA, Yu MC, Ross RK, et al. The CAG and GGC Microsatellites of the Androgen Receptor Gene Are in Linkage Disequilibrium in Men with Prostate Cancer. Cancer Res 1995;55:1937-40. [PubMed]
- Hardy DO, Scher HI, Bogenreider T, et al. Androgen Receptor CAG Repeat Lengths in Prostate Cancer: Correlation with Age of Onset. J Clin Endocrinol Metab 1996;81:4400-5. [PubMed]
- Giovannucci E, Stampfer MJ, Krithivas K, et al. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci U S A 1997;94:3320-3. [Crossref] [PubMed]
- Sartor O, Zheng Q, Eastham JA. Androgen receptor gene CAG repeat length varies in a race-specific fashion in men without prostate cancer. Urology 1999;53:378-80. [Crossref] [PubMed]
- Edwards SM, Badzioch MD, Minter R, et al. Androgen Receptor Polymorphisms: Association with Prostate Cancer Risk, Relapse and Overall Survival. Int J Cancer 1999;84:458-65. [Crossref] [PubMed]
- Bratt O, Borg A, Kristoffersson U, et al. CAG repeat length in the androgen receptor gene is related to age at diagnosis of prostate cancer and response to endocrine therapy, but not to prostate cancer risk. Br J Cancer 1999;81:672-6. [Crossref] [PubMed]
- Chamberlain NL, Driver ED, Miesfeld RL. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res 1994;22:3181-6. [Crossref] [PubMed]
- Luisi BF, Xu WX, Otwinowski Z, et al. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature 1991;352:497-505. [Crossref] [PubMed]
- Dahlman-Wright K, Wright A, Gustafsson JA, et al. Interaction of the Glucocorticoid Receptor DNA-binding Domain with DNA as a Dimer is Mediated by a Short Segment of Five Amino Acids. J Biol Chem 1991;266:3107-12. [PubMed]
- Clinckemalie L, Vanderschueren D, Boonen S, et al. The hinge region in androgen receptor control. Mol Cell Endocrinol 2012;358:1-8. [Crossref] [PubMed]
- Robinson D, Van Allen EM, Wu YM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015;161:1215-28. [Crossref] [PubMed]
- Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations in Cancer. Nucleic Acids Res 2019;47:D941-7. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Steketee K, Timmerman L, Ziel-van der Made AC, et al. Broadened ligand responsiveness of androgen receptor mutants obtained by random amino acid substitution of H874 and mutation hotspot 877 in prostate cancer. Int J Cancer 2002;100:309-17. [Crossref] [PubMed]
- Tan J, Sharief Y, Hamil KG, et al. Dehydroepiandrosterone Activates Mutant Androgen Receptors Expressed in the Androgen-Dependent Human Prostate Cancer Xenograph CWR22 and LNCaP cells. Mol Endocrinol 1997;11:450-9. [Crossref] [PubMed]
- Hara T, Miyazaki J, Araki H, et al. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Res 2003;63:149-53. [PubMed]
- Schalken J, Fitzpatrick JM. Enzalutamide: targeting the androgen signalling pathway in metastatic castration-resistant prostate cancer. BJU Int 2016;117:215-25. [Crossref] [PubMed]
- Korpal M, Korn JM, Gao X, et al. An F876L Mutation in Androgen Receptor Confers Genetic and Phenotypic Resistance to MDV3100 (Enzalutamide). Cancer Discov 2013;3:1030-43. [Crossref] [PubMed]
- Joseph JD, Lu N, Qian J, et al. A Clinically Relevant Androgen Receptor Mutation Confers Resistance to Second-Generation Antiandrogens Enzalutamide and ARN-509. Cancer Discov 2013;3:1020-9. [Crossref] [PubMed]
- Auchus RJ, Yu MK, Nguyen S, et al. Use of prednisolone with abiraterone acetate in metastatic castration-resistant prostate cancer. Oncologist 2014;19:1231-40. [Crossref] [PubMed]
- Rehman Y, Rosenberg JE. Abiraterone acetate: oral androgen biosynthesis inhibitor for treatment of castration-resistant prostate cancer. Drug Des Devel Ther 2012;6:13-8. [Crossref] [PubMed]
- Steinman RM, Cohn ZA. Identification of a Novel Cell Type in Peripheral Lymphoid Organs of Mice. J Exp Med 1973;137:1142-62. [Crossref] [PubMed]
- NobelPrize.org. Nobel Prize in Medicine and Physiology 1988, Nobel Media AB 2019 [online]. Available online: [accessed 25th January 2019].https://www.nobelprize.org/prizes/medicine/1988/summary/
- Di Lorenzo G, Buonerba C, Kantoff PW. Immunotherapy for the treatment of prostate cancer. Nat Rev Clin Oncol 2011;8:551-61. [Crossref] [PubMed]
- Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel-T Immunotherapy for Castration-Resistant Prostate Cancer. N Engl J Med 2010;363:411-22. [Crossref] [PubMed]
- Anassi E, Ndefo UA. Sipuleucel-T (Provenge) Injection The First Immunotherapy Agent (Vaccine) For Hormone-Refractory Prostate Cancer. P T 2011;36:197-202. [PubMed]
- NobelPrize.org, Nobel Prize in Medicine and Physiology 2011, Nobel Media AB 2019, [online]. Available online: [accessed 25th January 2019].https://www.nobelprize.org/prizes/medicine/2011/summary/
- Fitzpatrick JM, de Wit R. Taxane mechanisms of action: potential implications for treatment sequencing in metastatic castration-resistant prostate cancer. Eur Urol 2014;65:1198-204. [Crossref] [PubMed]
- Sweeney CJ, Chen YH, Carducci M, et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. N Engl J Med 2015;373:737-46. [Crossref] [PubMed]
- James ND, Sydes MR, Clarke NW, et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormonal therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised control trial. Lancet 2016;387:1163-77. [Crossref] [PubMed]
- Goodall J, Mateo J, Yuan W, et al. Circulating Cell-Free DNA to Guide Prostate Cancer Treatment with PARP Inhibition. Cancer Discov 2017;7:1006-17. [Crossref] [PubMed]


