Decoding the androgen receptor splice variants
Introduction
Prostate cancer is a common cause of cancer-related death in aging males in the Western world. Because hormone therapy is the standard of care for men with metastatic prostate cancer, most men succumb to the disease following developing resistance to at least one of the hormone therapy regimens. Castration-resistant prostate cancer (CRPC) is a term used to describe prostate cancers that relapse following first-line hormone therapy (1). It is known that CRPC is not completely refractory to further hormonal manipulation, and AR signaling remains as a pivotal driver for disease progression despite castrate levels of androgens (2,3). Sustained AR signaling may be mediated by a number of mechanisms, including AR gene amplification and overexpression (4-6), intra-tumoral androgen synthesis (7), overexpression of AR coactivators (8), aberrant kinase pathway activation (9-11), AR mutation (12), and constitutively active AR splice variants (13).
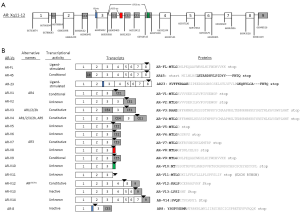
Novel therapies have been recently developed to treat CRPC patients by targeting overexpressed AR and intra-tumoral androgen synthesis. Abiraterone acetate, designed to inhibit CYP17A1, was approved on November 2011 for treating metastatic CRPC previously treated with docetaxol (14), with expanded indication approved on December 2012 to include patients who did not receive prior docetaxol (15). Enzalutamide, a more potent anti-androgen, was approved on August 2012 for post-docetaxol metastatic CRPC (16). The successful clinical development of these two new agents (14-16) underscores the importance of understanding the mechanism of sustained AR signaling in CRPC. In this light, most AR splice variants identified so far do not contain the intended therapeutic target, the AR ligand-binding domain (LBD), for any of the existing hormone therapy regimens including the two new agents. In this review, we will discuss discovery and characterization of the structural and functional diversity of AR splice variants for which the key features have been documented in the literature (key features of the 18 AR splice variants are summarized in Figure 1), their potential roles in mediating constitutively active AR signaling, and key areas of investigation to establish them as a mechanism of CRPC, particularly in the setting of resistance to abiraterone and enzalutamide.
The canonical AR-FL
In a normal male genome, there is only one copy of the AR gene located on Xq11-12. The AR gene is considered the most important gene in prostate cancer. The AR-FL cDNA was first cloned in 1988 (17). Structurally AR-FL resembles other nuclear receptors, containing a highly conserved DNA binding domain (DBD) encoded by Exon 2/3, a ligand binding domain (LBD) encoded by Exon 4-8 at C-terminus with lower sequence homology, a poorly conserved N-terminal domain (NTD) encoded by Exon 1, as well as a hinge region encoded by Exon 3/4 (18). Unique to AR, it has a long NTD domain (~538 amino acids) harboring two transactivating regions, termed transcription activation unit 1 (TAU1) and 5 (TAU5), that are indispensible for AR activation (19).
Earlier reports of AR splice variants: AR45 and AR23
In 2005, a NTD-truncated AR isoform with a deduced molecular weight around 45 kDa (so called AR45) was discovered by 5' rapid amplification of cDNA ends (5' RACE) from human placenta RNA (20). AR45 contains an intact DBD, hinge region, LBD, and a novel seven amino-acid long N-terminal peptide encoded by the novel exon 1b located ~22.1 kb downstream of AR exon 1 (Figure 1). The AR45 mRNA was found mainly in heart but also detected in skeletal muscle, uterus, prostate, breast, and lung (20). Exogeneously expressed AR45 did not stimulate the transactivation of androgen response element (ARE)-luciferase reporter in the presence of ligand. AR45 was proposed as a dominant negative AR that suppresses the function of AR-FL (20). In 2007, another AR splice variant, named AR23, was identified from a CRPC bone metastasis specimen (21). AR23 resulted from aberrant splicing of a 69 bp intron 2 sequence (corresponding to 23-amino acid residues), leading to in-frame insertion of a 23-amino acid sequence between two zinc fingers of the DBD (Figure 1). The genomic function of AR23 was not established because it does not translocate to the nucleus upon ligand binding, though cytoplasmic AR23 was partially active in androgen-responsive promoter reporter assays (21).
AR Splice variants lacking LBD due to splicing of cryptic exons
AR splice variants drew more attention since 2008 primarily due to the discovery of a number of variants that lack LBD. Such variants have the potential to mediate constitutively active AR function, on the basis of earlier in vitro studies on AR deletion constructs generated in the laboratory (22). In 2008, Dehm et al. performed 3' RACE with primers anchored at exon 1 and identified a new exon (termed exon 2b) 37 kb downstream of exon 2, in the CWR22Rv1 cell line that demonstrated ligand-independent AR activity (23). Splicing of exon 2b yielded two novel C-terminally truncated AR variants, AR1/2/2b and AR1/2/3/2b (23). Due to the presence of stop codons in exon 2b, LBD was replaced by the variant-specific 11-aa peptide encoded by exon 2b. Both variants demonstrated constitutively active AR function by in vitro luciferase reporter assays. AR1/2/2b also lacks the second zinc finger of the DBD, while AR1/2/3/2b retains the entire DBD. Because the transcript structure of AR1/2/3/2b was explained by a duplicated DNA sequence unique to CWR22Rv1 cells (24), this variant was thought to be specific to this cell line. In contrast, AR1/2/2b was more commonly found in other PCa cell lines, including VCaP, LNCaP, and LAPC4, as well as PCa xenografts (23).
In 2009, Hu et al. reported the identification of more AR cryptic exons in both cell lines and clinical specimens (25). Using a strategy combining exhaustive analysis of expressed sequence tags mapped to the human AR locus and experimental cloning to determine the precise splice junctions, Hu et al. identified three cryptic exons named CE1, CE2, and CE3 in intron 3, and CE4, identical to exon 2b (23) discovered by Dehm et al. (25). Splicing of the cryptic exons generated seven AR splice variants (named AR-V1 to AR-V7) (Figure 1), all lacking LBD due to stop codons present in the transcribed “intronic” sequences (i.e., cryptic exons). Among these, AR-V1 and AR-V7 were readily detectable in clinical prostate cancer specimens, with ~20-fold higher levels detected in CRPC specimens compared to hormone naïve prostate tumors. Importantly, a variant-specific antibody was developed for AR-V7, and used to detect the translated product of AR-V7 in prostate cancer cell lines and xenografts. Both PSA reporter assays and expression microarray analysis confirmed that AR-V7 was constitutively active in driving expression of canonical androgen-responsive genes (e.g., KLK3, KLK2, and NKX3.1) in an androgen-independent manner (25).
Guo et al. reported the discovery of LBD-truncated variants AR3, AR4, and AR5 using 3' RACE in 2009 (26). AR3, AR4, and AR5 contained coding sequences identical to those in AR-V7, AR-V1, and AR-V4, respectively. A variant-specific polyclonal antibody was also developed for AR3 (AR-V7), and used to detect protein expression in both hormone naïve and CRPC specimens. In addition, knockdown of AR3 in CWR22Rv1/CWR-R1 cells revealed a set of 117 genes that were preferentially regulated by AR3 (26). This study also reported the cloning of multiple additional variants that were not further characterized.
AR splice variants discovered by other approaches
Combining 3' RACE with next generation sequencing, Watson et al. not only confirmed the known AR-V1 and AR-V7, but also found 4 more AR splice variants named AR-V8 to AR-V11 (27) (Figure 1). This experiment was carried out in the VCaP cells, a prostate cancer cell line derived from vertebral metastatic lesion of a CRPC patient (27,28) that was shown to express the AR-V7 protein (25). These four new AR variants show splicing junctions between exon 3 and different regions of intron 3, with the predicted AR variant proteins truncated after AR DBD with 10-39 amino acid extension before the stop codon (Figure 1). Using VCaP xenograft in SCID mice, Watson et al. found that AR-Vs (AR-V7 and AR-V1) and AR-FL were upregulated by castration in both mRNA and protein levels, while re-administration of testosterone suppressed the expression of both AR-FL and AR-V7 in VCaP cells. Similar regulation of AR-FL and AR-Vs by androgens was also demonstrated in LuCaP35 xenografts with modest variation. However, only AR-V7, but not AR-V1, conferred gain-of-function on accelerating the LNCaP xenograft growth in castrated mice and colony formation in soft-agar assay (27).
More recently, Hu et al. employed a modified RNA amplification method, termed selective linear amplification of sense RNA (SLASR), for unbiased detection of transcribed AR sequences using arrayed 60-mer probes tiled across the human AR gene locus, directly in clinical CRPC specimens (29). This study provided a snapshot of the expression peaks along genomic sequences downstream of AR exon 3 and identified 3 new variants named AR-V12 to AR-V14 (Figure 1). Importantly, this study revealed expression peaks within intron 3 as well as sequences further downstream of exon 8 (named exon 9). These previously unappreciated expressed sequences have the potential to participate in AR splicing. One example is AR-V12 (Figure 1), which has the same open reading frame with ARv567es (see below) but contained untranslated sequences mapped to exon 9.
ARv567es and AR8
Sun et al. investigated the AR isoforms in a panel of 25 LuCaP prostate cancer xenografts (30). With a primer set anchored on exons 2 and 8 in RT-PCR, a short AR transcript spanning exon 2 to 8 was discovered. Sequencing revealed a novel AR variant arising from skipping of exons 5 to 7 while retaining the full sequence of exons 1 to 4 and exon 8. This new variant is named ARv567es (30). This exon combination (1/2/3/4/8) shifts the open reading frame (ORF) of ARv567es to an early stop codon just after the first 29 nucleotides of exon 8 (Figure 1). ARv567es is unique in that it retains the full hinge domain encoded by part of exons 3 and 4. The AR hinge domain contained important sequences for AR localization and activity (31). Similar to AR-V7, ARv567es activates androgen-responsive genes (such as KLK3, TMPRSS2, and NKX3.1) in a hormone-independent manner when ectopically expressed in LNCaP cells (30). The coding sequence for ARv567es is identical to AR-V12, which is encoded by a transcript containing exons 1/2/3/4/8/9 as later reported in Hu et al. (29) (Figure 1). However, more in-depth studies of ARv567es have been hampered by lack of a variant-specific antibody, as well as lack of suitable sequences to target for variant-specific knockdown that is an important tool to determine protein translation and function.
In CWR-R1 cells, Yang et al. identified a membrane-associated AR variant, named AR8 by RACE (32). Using an alternative splicing acceptor site 186 bp upstream of exon 3, the deduced protein of AR8 (Exon1-3'-3-3b/CE3) contained 33 unique amino acids after the NTD domain. Higher expression level of AR8 was detected in castration-resistant cell lines (C4-2, C4-2B, CWR22Rv1). This C-terminal truncated AR-V has no DBD or LBD and no transactivating function in ARE-luciferace reporter assay. Possibly due to palmitoylation of two cysteine residues within its unique C-terminal sequence, this protein was found mainly in plasma membrane when overexpressed in COS-1 cells or PCa cells (LNCaP and CWR-R1). Membrane-bound AR8 complexes with AR-FL and EGFR and may serve as a mediator in Src-induced AR activation (32).
Nuclear localization of AR splice variants
A prerequisite for AR to exert its genomic function is to enter the nucleus. Upon androgen binding to LBD, AR-FL exposes a nuclear localization signal (NLS) within C-terminal end (CTE) of DBD and hinge region to interact with importin proteins for translocation through the nuclear pore complex (33). A canonical bipartite nuclear localization signal (NLS) was mapped at the junction of DBD and the hinge region (amino acid 617-RKCYEAGMTLG--ARKLKK-633) (34). With the exception of AR-12/ARv567es, other AR isoforms may have variable capability in nuclear import due to loss of NLS. Evidence provided by immunofluorescent staining supports constitutive nuclear localization of AR-V7/AR3 and AR-12/ARv567es in the absence of androgens, while AR-V1, AR-V9, and AR-V13 are mainly cytoplasmic (25,27,29,30), possibly due to lack of basic amino acids characteristic of the bipartite nuclear localization sequence (NLS) (35). Interestingly, genomic functions of AR-Vs do not always parallel to their localization. For example, AR-V1 and AR-V9 showed ligand-independent activity in LNCaP cells but not in PC-3 cells (29). Such variants were termed “conditionally active” variants (29), to differentiate them from constitutively active variants (see below), because their functions are conditional on the cellular context. To further understand the nuclear transport of AR-V7 and its relation with AR-V transcriptional function, Chan et al. showed that part of its unique sequence at C-terminus (aa 628-EKFRVGNCKHLKMTRP-643) resembles the truncated bipartite AR NLS. Mutation of amino acid residues K629 and R631 to alanine in AR-V7 shifted its expression from predominantly nuclear to a mixed nuclear/cytoplasmic pattern; while alanine mutation at K636 or K639 had no effect on nuclear localization of AR-V7 (36).
Diverse and cell-specific functions of AR splice variants
Among the AR-Vs listed in Figure 1, AR-V7 (also named AR3) and ARv567es have received more attention due to their unequivocal constitutively active nuclear functions (25,26,30). Both AR-Vs activate transcription of canonical AR-FL target genes when overexpressed in cell lines with or without activated AR-FL. Other AR-Vs may be conditionally active, i.e., their transcriptional activities are cell type-specific (37). For example, AR-V1 and AR-V9 demonstrated transcriptional activity when introduced in AR-FL positive LNCaP cells but not in the AR-FL negative PC-3 cells (37). It is possible that the conditional activity of AR-V1 and AR-V9 may require nuclear localization that was not readily detected by immunofluorescence. Previous studies showed androgen receptor (AR) deletion mutants that retain a partially truncated LBD did not have constitutive activity (22,35,38). Hu et al. demonstrated examples of inactive AR splice variants that retain a partially truncated LBD, including AR-V13 and AR-V14 (37) (Figure 1).
Expression levels of AR-V7 are dramatically increased after suppression of AR-FL signaling by androgen depletion, AR-FL knockdown, or treatment with enzalutamide in VCaP cells and LNCaP95 cells but not in LNCaP and CWR22Rv1 cells, suggesting that in addition to cell-context specific functions of AR splice variants, the regulation of AR variant levels may also depend on a specific cellular context (39).
Molecular origin of AR splice variants
In clinical specimens, AR splice variants coexist with AR-FL, and the expression levels of individual AR variants almost always constitute a small fraction of the expression level of AR-FL (25,27). In addition, AR splice variants are also expressed in benign prostate epithelium (25,30), again at a much lower level relative to AR-FL. AR-FL is often overexpressed in CRPC due to AR gene amplification (40,41) or other genomic changes (42). In addition, elevated AR expression in CRPC may involve AR self regulation. Cai et al. showed that lysine-specific demethylation-1 (LSD-1) was recruited to AREs in intron 2 of the AR gene and acts as a repressor when AR-FL was activated. This recruitment was abolished when androgen was depleted (43). It is therefore possible that expression of AR splice variants are generally coupled with the transcriptional output from the AR gene locus (44). Supporting this possibility, AR variant levels were downregulated by testosterone replacement in castrated mice in parallel with a decrease of AR-FL in VCaP and LuCaP35 xenografts (27). In cell line models (VCaP and LNCaP95) with higher levels of AR-FL and low levels of AR variants, suppression of AR-FL signaling by enzalutamide resulted in an unequivocal increase of AR-V7, and a relatively moderate increase of AR-FL (39). In addition, increased expression of AR-FL, AR-V7 and ARv567es was also observed in castration-resistant LuCaP xenograft (LuCaP23CR and LuCaP35CR) when AR-FL signaling was inhibited by abiraterone (45). Thus, although AR-V expression may not strictly parallel that of AR-FL, and the magnitude of AR-V mRNA increase is generally greater than that of AR-FL in CRPC and in experimental models (39), AR-V expression is strongly coupled with AR-FL expression.
In some cell line and xenograft models (e.g., CWR22 and LuCaP86.2), AR intragenic rearrangement or deletions may be responsible for high AR variant expression. In CWR22Rv1 cells, an intragenic copy number increase occurred in an approximately 35-kb AR genomic segment between introns 2 and 3, with the rearranged segment flanked by long interspersed nuclear element (5'-LINE-1 and 3'-LINE-1) (46). To further investigate the association of focal imbalance of the AR gene and AR variant expression, Li et al. demonstrated a complex pattern of focal copy number imbalance with or without AR gene amplification. In LuCaP 86.2 xenograft cells, an 8579-bp deletion of AR exons 5, 6, and 7 may be responsible for the high level of ARv567es variant expression (47). The extent to which similar genomic alterations contribute to the generation of AR splice variants in clinical specimens remains unclear.
Detection of AR splice variants in clinical specimens
The majority of AR-Vs listed in Figure 1 can be detected in prostate cancer tissue specimens by RT-PCR (25-27,32,37). Alternatively spliced transcripts containing premature stop codons may be degraded through the nonsense-mediated decay (NMD) mechanism (48). Therefore it is also critical to demonstrate protein expression by detecting the corresponding variant protein product in order to draw functional relevance. Variant-specific antibodies have been reported for AR-V7 (AR3) (25,26), AR8 (49), and AR1/2/2b (50). In all these efforts, the variant-specific c-terminal peptides were used (Figure 1) as antigens. Among these, AR-V7/AR3 remains the only AR splice variant with a proven protein product that can be detected in clinical specimens using variant-specific antibodies (25,26,39), including a monoclonal antibody to AR-V7 (39). An alternative approach to detect the potential existence of LBD-truncated AR variants is to combine data obtained using antibodies recognizing AR-NTD and AR-LBD, respectively. For example, Zhang et al. showed a wide distribution of the AR-NTD/LBD ratio in clinical CRPC specimens (51). Higher ratios of NTD/LBD were detected in more aggressive tumors. However, this approach is based on the assumption that excess AR-NTD detected in CRPC specimens originated from the expression of AR-Vs.
Genomic functions of AR splice variants
A key question in relation to the genomic functions of AR splice variants is whether they active the same transcriptional programs directed by AR-FL. Hu et al. showed data suggesting that endogenously induced AR variants are not sufficient to “rescue” the suppressed AR-FL, when a set of canonical AR-FL target genes are evaluated (39). Instead, increased expression of AR variant paralleled the increased expression of cell cycle genes, and forced expression of both AR-V7 and ARv567ES induced the same set of cell cycle genes in both the presence of absence of canonical AR-FL signaling (39). Li et al. performed gene expression profiling in rearrangement-driven AR-V positive cells following specific knockdown of the AR-FL and AR-Vs to differentiate genes activated by the two different receptor molecules (52). AR-V-dependent cell cycle genes were found to demonstrate a biphasic response. They were induced at low AR-V levels but repressed when higher AR-Vs were expressed in the cells. This observation mirrors the canonical biphasic androgen-stimulated (i.e., AR-FL-mediated) growth response observed in cell line models. The findings suggest that AR-V expression reactivates and restores the AR-FL transcriptional programs, rather than by targeting a unique set of genes. These seemingly opposing findings may be explained by cell-context differences as well as the different methodologies used in the studies. More in-depth analysis will address cell-context specific genomic functions mediated by the AR splice variants.
Future directions and priorities
In spite of intense interest in the putative role of AR splice variants in CRPC and the years that have elapsed since their discovery and characterization, the field is still in infancy and investigations encompassing the full spectrum of mechanistic characterization and clinical translation are still at a nascent stage. Successful clinical development of abiraterone and enzalutamide (14-16), both intended to target the AR LBD (which is missing in AR-Vs), is directly driven by laboratory mechanistic studies establishing intra-tumoral androgens and AR protein overexpression as the key molecular determinants of CRPC (2). Thus we envision that mechanistic studies dissecting the genomic functions of different AR molecules will facilitate efforts in developing new therapies to overcome resistance to abiraterone and enzalutamide. Given the expanded clinical use of abiraterone and enzalutamide, there is an urgent need to dissect the various putative mechanisms of resistance to these new, more potent inhibitors of AR-FL signaling. Although AR splice variants provide a biologically plausible explanation for therapeutic resistance, the concept has not been validated in clinical specimens due to the recent approval of the two new agents, and consequently lack of sufficient number of relevant specimens collected from treated patients. Nevertheless, the discovery of AR splice variants has already stimulated efforts to develop novel agents that target all AR molecules to overcome resistance (53-64). Further conceptual advances in the field will provide a sustained impetus for such efforts.
Acknowledgements
The authors wish to thank all collaborators and investigators who contributed to the cited studies. Due to space limitations as well as the limited scope of the review article, many worthy studies are not cited in this article.
Funding: Some of the cited research work conducted in the authors’ laboratories were supported by the NIH/NCI Specialized Program in Research Excellence (SPORE) in Prostate Cancer grant P50 CA058236 (PI: William Nelson), the Patrick C. Walsh Prostate Cancer Research Foundation (JL), and the Prostate Cancer Foundation (PI: Johann de Bono).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Scher HI, Buchanan G, Gerald W, et al. Targeting the androgen receptor: improving outcomes for castration-resistant prostate cancer. Endocr Relat Cancer 2004;11:459-76. [PubMed]
- Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol 2008;8:440-8. [PubMed]
- Attard G, Cooper CS, de Bono JS. Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell 2009;16:458-62. [PubMed]
- Koivisto P, Kononen J, Palmberg C, et al. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res 1997;57:314-9. [PubMed]
- Visakorpi T, Hyytinen E, Koivisto P, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet 1995;9:401-6. [PubMed]
- Bubendorf L, Kononen J, Koivisto P, et al. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res 1999;59:803-6. [PubMed]
- Cai C, Balk SP. Intratumoral androgen biosynthesis in prostate cancer pathogenesis and response to therapy. Endocr Relat Cancer 2011;18:R175-82. [PubMed]
- Gregory CW, He B, Johnson RT, et al. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res 2001;61:4315-9. [PubMed]
- Signoretti S, Montironi R, Manola J, et al. Her-2-neu expression and progression toward androgen independence in human prostate cancer. J Natl Cancer Inst 2000;92:1918-25. [PubMed]
- Culig Z, Hobisch A, Cronauer MV, et al. Androgen receptor activation in prostatic tumor cell lines by insulin-like growth factor-I, keratinocyte growth factor, and epidermal growth factor. Cancer Res 1994;54:5474-8. [PubMed]
- Gao H, Ouyang X, Banach-Petrosky WA, et al. Combinatorial activities of Akt and B-Raf/Erk signaling in a mouse model of androgen-independent prostate cancer. Proc Natl Acad Sci U S A 2006;103:14477-82. [PubMed]
- Linja MJ, Visakorpi T. Alterations of androgen receptor in prostate cancer. J Steroid Biochem Mol Biol 2004;92:255-64. [PubMed]
- Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer 2011;18:R183-96. [PubMed]
- de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011;364:1995-2005. [PubMed]
- Ryan CJ, Smith MR, de Bono JS, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013;368:138-48. [PubMed]
- Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367:1187-97. [PubMed]
- Chang CS, Kokontis J, Liao ST. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science 1988;240:324-6. [PubMed]
- Hu R, Denmeade SR, Luo J. Molecular processes leading to aberrant androgen receptor signaling and castration resistance in prostate cancer. Expert Rev Endocrinol Metab 2010;5:753-64. [PubMed]
- Jenster G, van der Korput HA, Trapman J, et al. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem 1995;270:7341-6. [PubMed]
- Ahrens-Fath I, Politz O, Geserick C, et al. Androgen receptor function is modulated by the tissue-specific AR45 variant. FEBS J 2005;272:74-84. [PubMed]
- Jagla M, Fève M, Kessler P, et al. A splicing variant of the androgen receptor detected in a metastatic prostate cancer exhibits exclusively cytoplasmic actions. Endocrinology 2007;148:4334-43. [PubMed]
- Jenster G, van der Korput HA, van Vroonhoven C, et al. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol 1991;5:1396-404. [PubMed]
- Dehm SM, Schmidt LJ, Heemers HV, et al. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res 2008;68:5469-77. [PubMed]
- Tepper CG, Boucher DL, Ryan PE, et al. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res 2002;62:6606-14. [PubMed]
- Hu R, Dunn TA, Wei S, Isharwal S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 2009;69:16-22. [PubMed]
- Guo Z, Yang X, Sun F, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res 2009;69:2305-13. [PubMed]
- Watson PA, Chen YF, Balbas MD, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A 2010;107:16759-65. [PubMed]
- Korenchuk S, Lehr JE, MClean L, et al. VCaP, a cell-based model system of human prostate cancer. In Vivo 2001;15:163-8.
- Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate 2011;71:1656-67. [PubMed]
- Sun S, Sprenger CC, Vessella RL, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest 2010;120:2715-30. [PubMed]
- Clinckemalie L, Vanderschueren D, Boonen S, et al. The hinge region in androgen receptor control. Mol Cell Endocrinol 2012;358:1-8. [PubMed]
- Yang X, Guo Z, Sun F, et al. Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J Biol Chem 2011;286:36152-60. [PubMed]
- Cutress ML, Whitaker HC, Mills IG, et al. Structural basis for the nuclear import of the human androgen receptor. J Cell Sci 2008;121:957-68. [PubMed]
- Saporita AJ, Zhang Q, Navai N, et al. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem 2003;278:41998-2005. [PubMed]
- Zhou ZX, Sar M, Simental JA, et al. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem 1994;269:13115-23. [PubMed]
- Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem 2012;287:19736-49. [PubMed]
- Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate 2011;71:1656-67. [PubMed]
- Jenster G, van der Korput JA, Trapman J, et al. Functional domains of the human androgen receptor. J Steroid Biochem Mol Biol 1992;41:671-5. [PubMed]
- Hu R, Lu C, Mostaghel EA, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 2012;72:3457-62. [PubMed]
- Edwards J, Krishna NS, Grigor KM, et al. Androgen receptor gene amplification and protein expression in hormone refractory prostate cancer. Br J Cancer 2003;89:552-6. [PubMed]
- Linja MJ, Savinainen KJ, Saramäki OR, et al. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res 2001;61:3550-5. [PubMed]
- Sharma A, Yeow WS, Ertel A, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest 2010;120:4478-92. [PubMed]
- Cai C, He HH, Chen S, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 2011;20:457-71. [PubMed]
- Liu LL, Xie N, Sun S, et al. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 2014;33:3140-50. [PubMed]
- Mostaghel EA, Marck BT, Plymate SR, et al. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin Cancer Res 2011;17:5913-25. [PubMed]
- Li Y, Alsagabi M, Fan D, et al. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res 2011;71:2108-17. [PubMed]
- Li Y, Hwang TH, Oseth LA, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene 2012;31:4759-67. [PubMed]
- Pan Q, Saltzman AL, Kim YK, et al. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev 2006;20:153-8. [PubMed]
- Yang X, Guo Z, Sun F, et al. Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J Biol Chem 2011;286:36152-60. [PubMed]
- Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem 2012;287:19736-49. [PubMed]
- Zhang X, Morrissey C, Sun S, et al. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PLoS One 2011;6:e27970. [PubMed]
- Li Y, Chan SC, Brand LJ, et al. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013;73:483-9. [PubMed]
- Andersen RJ, Mawji NR, Wang J, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 2010;17:535-46. [PubMed]
- Sadar MD. Small molecule inhibitors targeting the “achilles’ heel” of androgen receptor activity. Cancer Res 2011;71:1208-13. [PubMed]
- Schmidt LJ, Tindall DJ. Androgen receptor: past, present and future. Curr Drug Targets 2013;14:401-7. [PubMed]
- Lim AC, Attard G. Improved therapeutic targeting of the androgen receptor: rational drug design improves survival in castration-resistant prostate cancer. Curr Drug Targets 2013;14:408-19. [PubMed]
- Levine PM, Imberg K, Garabedian MJ, et al. Multivalent peptidomimetic conjugates: a versatile platform for modulating androgen receptor activity. J Am Chem Soc 2012;134:6912-5. [PubMed]
- Yamashita S, Lai KP, Chuang KL, et al. ASC-J9 suppresses castration-resistant prostate cancer growth through degradation of full-length and splice variant androgen receptors. Neoplasia 2012;14:74-83. [PubMed]
- Nakka M, Agoulnik IU, Weigel NL. Targeted disruption of the p160 coactivator interface of androgen receptor (AR) selectively inhibits AR activity in both androgen-dependent and castration-resistant AR-expressing prostate cancer cells. Int J Biochem Cell Biol 2013;45:763-72. [PubMed]
- Zengerling F, Streicher W, Schrader AJ, et al. Effects of sorafenib on C-terminally truncated androgen receptor variants in human prostate cancer cells. Int J Mol Sci 2012;13:11530-42. [PubMed]
- Oh SJ, Erb HH, Hobisch A, et al. Sorafenib decreases proliferation and induces apoptosis of prostate cancer cells by inhibition of the androgen receptor and Akt signaling pathways. Endocr Relat Cancer 2012;19:305-19. [PubMed]
- Mashima T, Okabe S, Seimiya H. Pharmacological targeting of constitutively active truncated androgen receptor by nigericin and suppression of hormone-refractory prostate cancer cell growth. Mol Pharmacol 2010;78:846-54. [PubMed]
- Cao B, Liu X, Li J, Liu S, et al. 20(S)-protopanaxadiol-aglycone downregulation of the full-length and splice variants of androgen receptor. Int J Cancer 2013;132:1277-87. [PubMed]
- Knudsen KE, Kelly WK. Outsmarting androgen receptor: creative approaches for targeting aberrant androgen signaling in advanced prostate cancer. Expert Rev Endocrinol Metab 2011;6:483-93. [PubMed]


