Entering an era of radiogenomics in prostate cancer risk stratification
Introduction
While it is clear that prostate cancer screening reduces mortality, it has led to an over detection and treatment of prostate cancer (1,2). Active surveillance (AS) has emerged as a safe alternative to immediate therapy. However, criteria for AS eligibility are primarily based on biopsy findings, which are inherently known to under sample the gland and potentially underestimate the grade and extent of cancer (3). As a result, there is always some uncertainty that a more aggressive tumor may have been missed. Given this limitation, better tools to enhance risk stratification are desperately needed.
Over the years, we have seen an increase in the role of multi-parametric magnetic resonance imaging (mpMRI) of the prostate in various aspects of prostate cancer detection and management (4-6). In addition, we have seen the emergence of genomic signatures that provide prognostic information to facilitate decision-making in prostate cancer (7,8). In fact, both these tools have been increasingly used to help decide on the need for treatment in men with newly diagnosed prostate cancer. While they both have good evidence supporting their role in risk stratification, they both have limitations. We believe by integrating these modalities we can enhance their individual performance characteristics and perhaps overcome some of their limitations. This concept of combining imaging, like mpMRI, with genomics is the tip of the iceberg of a rapidly developing and promising field known as “radiogenomics”.
Radiomics is an emerging field, and involves a process of extracting quantitative data on imaging characteristics such as texture, shape, and other features that are linked to the presence of cancer and its aggressiveness (9). Given the trove of quantifiable information available in mpMRI, it lends itself nicely to the study of radiomics in prostate cancer. At its most simple form, radiogenomics in prostate cancer could refer to the integration of genomic testing and mpMRI, two modalities that are already in rapid use in prostate cancer risk stratification. While we see much promise for these tools in many aspects of prostate cancer, their greatest potential utility is in the selection of men for AS versus immediate treatment.
As a result, we have framed this review and the discussion of radiogenomics and their individual components as it relates to risk stratification in AS. We begin with a short discussion on AS, followed by some of the evidence and limitations behind mpMRI and genomic testing in this space, and the rationale behind their integration. We follow with an introduction to an ongoing prospective, interventional trial at the University of Miami, where we are investigating the individual and combined performance of these two tests. Finally, we will conclude with a discussion of radiogenomics at a more granular level, which requires further validation and while not ready for clinical use, holds much promise to improve risk stratification and decision-making in prostate cancer.
AS for prostate cancer
The principle objective of AS is to reduce the overtreatment of indolent prostate cancer. This concept is supported by trials comparing immediate therapy to observation that have shown no difference in overall or prostate cancer specific mortality (10,11). However, patients with intermediate and high-risk prostate cancer fared better with immediate treatment, suggesting that patient selection is crucial to outcome. Over the years, we have seen a rise in the utilization of AS, with reports from the CaPSURE, MUSIC Collaboration, and other registries showing a global adoption of AS in low risk patients, with rates as high at 74% and 91%, in low and very low risk patients, respectively, in Sweden (12-16). Even among patients there has been a considerable increase in the acceptance of AS (13).
Typically, selection criteria for AS is conservative and restricted to patients with low volumes of indolent or low risk cancer (Grade Group 1) (3). Numerous studies have shown in carefully selected patients like this, long term cancer specific survival rates are near perfect, with very few men dying from prostate cancer (3). This, combined with level one evidence showing that AS is a safe alternative to immediate therapy, has resulted in the incorporation of AS into several national guidelines as a primary and recommended modality of treatment in men with low risk prostate cancer (17,18). Acknowledging its benefits, there is a debate whether AS protocols can be extended to patients who harbor low volume, intermediate risk prostate cancer. Cooperberg et al. has shown that a well selected cohort of men with intermediate risk prostate cancer can be considered for AS with similar short to intermediate risk outcome as men with lower risk disk (19). However, long term date from the University of Toronto, has suggested that men with intermediate risk prostate cancer are a greater risk of long term metastasis when selecting observation over immediate treatment (20). This study suggested the need for caution and careful selection when considering AS in men with intermediate risk prostate cancer. In fact, a recent study from John Hopkins University looked at 6,721 men who underwent radical prostatectomy and long term follow up and concluded that traditional clinical and pathological criteria are incapable of identifying a favorable subset of patients with intermediate risk prostate cancer who would have similar outcomes to low risk patients if placed on surveillance, suggesting the limitations of our clinical information and decision making based on it (21). We believe that the two main limitations hindering accurate risk stratification are tumor heterogeneity and multi-focality in prostate cancer. This can result in an underestimation of tumor grade and extent, and misguided decision-making. Even in low risk men, there are those found to have high-risk prostate cancer that was missed on initial sampling of the prostate (22). Tools such as mpMRI and genomics markers bring hope to improving risk stratification and provide better tools for selecting men who are appropriate for AS.
mpMRI and MRI targeted biopsy for prostate cancer detection and risk stratification
MRI (anatomical sequence T2) of the prostate was initially used after a diagnosis of prostate cancer to look for possible extracapsular. However, with the addition of functional sequences such as diffusion weighted imaging (DWI) and dynamic contrast enhancement (DCE), mpMRI of the prostate has emerged as one of the main tools for primary prostate cancer detection and risk stratification. While the primary role of mpMRI in AS would be to rule out the presence of a significant cancer that would compromise outcomes if it were observed, some the best evidence on its performance in this regard comes from the prostate cancer detection literature. One of the best studies supporting mpMRI is the PROMIS trial, which enrolled 575 men who undergoing an initial biopsy for evaluation of prostate cancer (4). All men underwent mpMRI of the prostate, followed by a TRUS guided extended template biopsy, and finally a transperineal template mapping biopsy, with cores spaced 5 mm apart, which served as the gold standard test for cancer detection. The mapping biopsy was the greatest strength of the trial as it provided the closest thing to having a radical prostatectomy on every man, reducing the concern for under sampling of the gland and providing a closer estimate of the true grade and extent of cancer in these men. The trial defined clinically significant cancer as Gleason score 4+3 or higher or a maximum cancer core length of ≥6 mm. The study found that mpMRI had a higher sensitivity (93% vs. 48%, P<0.0001) and a lower specificity (41% vs. 96%, P<0.0001) for the detection of clinically significant cancer. However, MRI missed clinically significant cancer 11% of the time using the above definition, and 24% of the time using a definition of any Gleason score 3+4 or higher cancer. As a result, the biggest concern with mpMRI is the worry about missed cancer, or false negatives, which is often represented by the negative predictive value (NPV), or the proportion of times a cancer was found in a person with a negative mpMRI. While this number can vary depending on the cohort and population being studied, a recent metanalysis by Moldovan et al. evaluated 48 studies and found the median NPV for any cancer, and significant cancer was 82.4%, and 88.1% respectively, with a range of 69.0% to 92.4% for any cancer, and 85.7% to 92.3% for significant cancer, respectively (6). The authors also found that the NPV of mpMRI was inversely related to the prevalence of prostate cancer with cohorts having a high prevalence of prostate cancer demonstrating a lower NPV. Additionally, a meta-analysis evaluating the performance of mpMRI in AS assessed 7 studies, encompassing a total of 1,028 men to look at the role of mpMRI in predicting reclassification at confirmatory biopsy (23). Three of the studies included only very low-risk patients, three included low-risk patients, and only one study allowed intermediate risk patients. The results showed in the AS patients, mpMRI had a pooled sensitivity and specificity of 69% (95% CI: 44–86%) and 78% (95% CI: 55–91%), respectively. The authors also reported that the NPV depended on the prevalence of cancer in the cohort. They summarized that for the typical AS patient entering observation, mpMRI that is suspicious for cancer resulted in a 3-fold increased risk of reclassification compared to a negative mpMRI, which resulted in a 60% lower risk.
Complementary to the emerging literature on mpMRI in prostate cancer, we have also seen a surge in the number of studies supporting MRI-based targeted biopsy of the prostate. There are a number of ways to perform an MRI directed biopsy including in-bore, fusion, and cognitive. However, the seminal study supporting the benefits of MRI guided biopsy comes from Siddiqui and colleagues at the NIH, who looked at 1,003 men who had a suspicious lesion on mpMRI and underwent MRI-US fusion and extended template biopsy of the prostate for evaluation of prostate cancer (24). The authors defined clinically significant prostate cancer as Gleason score 4+3 or higher cancer or high volume of Gleason score 3+4 cancer. The targeted biopsies resulted in a 30% increase in the detection of clinically significant cancers and a 17% reduction in the diagnosis of indolent cancers. The authors reported that adding template cores to targeted biopsy allowed for 22% more cancers to be diagnosed, but only 5% of these were clinically significant. They also reported that these findings were mainly driven by men with a previous negative biopsy. The authors found no significant difference in the distribution of cancers between targeted and template cores in men who had never undergone previous biopsy. In a similar study, Recabal et al. evaluated 206 men with low risk prostate cancer undergoing AS who underwent mpMRI and targeted and/or template biopsy of the prostate (25). The authors found that 34% of the cohort had a negative or low suspicion mpMRI and underwent systematic biopsy only, while 64% had at least one region of suspicion on mpMRI and underwent targeted and systematic biopsy. Results showed upgrading in 35% of the cohort, with 47% of men who had a suspicious mpMRI being found to have a more aggressive cancer within the prostate. However, its noteworthy that a reasonable proportion of clinically significant cancers were found only on the systematic biopsies (10–17%). This suggests that both targeted and systematic biopsy should be used for the optimal detection of clinically relevant cancer in men on AS.
While there is good evidence supporting the role of mpMRI and targeted biopsy in the selection of men for AS, there is less literature available on the role of mpMRI in monitoring for tumor progression. With respect to the selection of patients for AS a previous study looking at men with low risk cancer who underwent mpMRI and targeted confirmatory biopsy found that an extra 10% of the patients who were otherwise eligible for AS based on systemic biopsies, get reclassified by the MRI targeted cores (26). A similar study by Stamatakis et al. found a 29% reclassification rate and concluded the mpMRI was a useful tool for selecting appropriate patients for AS (27). To better quantify the amount of reclassification resulting from mpMRI and targeted biopsy we previously investigated a consecutive cohort of men undergoing fusion biopsy for evaluation of prostate cancer. We selected patients who were eligible for one of seven published AS criteria based on systematic biopsy cores alone. The addition of the MRI target cores resulted in an extra 10–40% of patients getting reclassified depending on the AS selection criteria utilized (28). We found that criteria with a minimum number of cores (usually two), resulted in the highest rates of reclassification most likely due to oversampling of a low risk tumor. To address this issue it is imperative that AS selection criteria be updated to reflect more contemporary evaluation and management strategies. For example, using a minimum percentage of cores positive instead of an absolute number resulted in much less reclassification due to only minimal increases in the volume of indolent disease (28). Unfortunately, as mentioned earlier the literature looking at the role of mpMRI in monitoring prostate cancer is sparse. There are a couple papers which have suggested that mpMRI can be helpful for detecting tumor progression, and help reduce the number of biopsies on AS. However, both of these studies had a limited number of patients and follow up to make any conclusive findings regarding the performance of mpMRI in monitoring cancer progression on AS (29,30).
Genomic markers in prostate cancer
In addition to mpMRI, we have seen innovation in the emergence of several genomic signatures that have been validated as independent predictors of adverse outcomes and are currently being used for decision-making in prostate cancer (8). Currently there are 4 commercially available genomic markers that can be performed on formalin fixed paraffin-embedded tissue from biopsy cores to provide prognostic information to aid in the decision to treat or observe prostate cancer (Genomic Health’s Oncotype Dx test®, Myriad’s Polaris test®, Genome Dx’s Decipher test®, and Metamark’s Promark®).
Genomic Health’s Oncotype Dx® test is a 17-gene signature that was specifically selected to address the issues of tumor heterogeneity and multi-focality (31). It was developed and validated in a multi-institutional study between the Cleveland Clinic and the University of California, San Francisco. The development of the signature began with 441 radical prostatectomy specimens, where the authors investigated 727 genes and filtered it down to 288 genes that predicted recurrence regardless of Gleason score. From these genes, they investigated 81 that had a biologically plausible mechanism of action in prostate cancer. Using biopsy specimens from 167 men, they were able to filter down to 58 genes that could be detected on small tissue samples from biopsy cores. From these 58 genes, a final 17 gene panel signature consisting of 12 genes from biologically distinct pathways and 5 reference genes were selected. The final signature was then validated in 395 men with matched biopsy and RP pathology, where it was found to predict the likelihood of adverse pathology at radical prostatectomy, which was defined as extracapsular extension (ECE) or primary pattern 4 prostate cancer.
Myriad Polaris® has a 31-gene cell cycle progression (CCP) signature that was originally developed to predict metastasis in a radical prostatectomy and watchful waiting cohort (32,33). The signature was validated for use in AS by Cooperberg et al. using 413 who underwent radical prostatectomy and had a minimum of 5 years of follow up available. The authors found that CCP scores were highly correlated with the likelihood of recurrence, and among both low and intermediate-high risk patients, the CCP score allowed a level of risk discrimination beyond that permitted by histopathology alone (34).
Genome Dx developed the Decipher Test®, which is a 22-gene panel signature that was also developed to predict metastasis using radical prostatectomy tissue (35,36). While it has been extensively validated, it has been investigated mostly for its primary use after radical prostatectomy to decide on the need for adjuvant radiotherapy or ADT (37). However, to assess its performance on biopsy samples, Klein et al investigated 57 men who had a biopsy and underwent radical prostatectomy and reported that the Decipher test on biopsy was the only preoperative independent predictor of prostate cancer recurrence (38).
Promark score® is based on an immunofluorescent assay analyzing 8 protein-based biomarkers from a formalin-fixed paraffin-embedded (FFPE) prostate biopsy sample (39). Clinical validation study conducted with 276 patients evaluated two co-primary end points: (I) to determine whether the 8-biomarker derived score predicts “favorable” pathology (radical prostatectomy Gleason score ≤3+4 and organ confined disease); (II) whether the assay score predicts radical prostatectomy Gleason score 3+3 disease (40). The AUC for determining “favorable” pathology by the assay score was 0.68 (P<0.001) and for determining radical prostatectomy Gleason score 3+3 disease was 0.65 (P<0.001). Furthermore, addition of the biomarker assay to NCCN (0.75 vs. 0.69) and D’Amico (0.75 vs. 0.65) risk groups yielded a better AUC when compared to the AUC’s of each of the classifications alone for determining “favorable” pathology.
Therefore, each of these signatures has good evidence validating itself as an independent predictor of prostate cancer outcomes and can provide us helpful information that we may not get from the histopathology. Unfortunately, none of these tests have been compared to each other with regards to their performance in AS. As a result, providers and patients remain in the dark about which of these signatures is the most reliable, with each company boosting their marker as superior over the others.
Additional benefits of utilizing biomarkers in clinical decision-making process are noted on the economic front. A study which utilized Genomic Prostate Score® (GPS) testing prospectively in clinical decision making on very low and low risk patients showed an increased utilization of AS and a net average saving of $2,286 per patient (41). However, these results need further validation in long term prospective, randomized trials to elucidate the true cost and benefit of using these tests in prostate cancer decision-making.
Prostate cancer heterogeneity and concern for genomics
While performing tests on radical prostatectomy tissue, the entire prostate is available and the tumor with the highest grade is often selected for genomic analysis. However, when the test is being performed on biopsy tissue, it is uncertain how vulnerable it is to the tumor heterogeneity and multi-focality issues that limit proper risk stratification. A recent study by Wei et al. looked at 4 radical prostatectomy specimens and conducted random biopsy sampling of the gland after it was removed. Each of the cores were sent for whole genome sequencing that specifically evaluated the expression of the individual genes involved in each of the signatures (42). They found that the expression levels of these genes were variable throughout the different biopsy cores, suggesting that different biopsy cores would yield different genomic results. This supports the concern that tumor heterogeneity is not only an issue for pathologic grading, but also for genomics.
Additionally, a recent abstract at the Genitourinary section of the American Society of Clinical Oncology annual meeting in 2018 investigated this concern specific to men with low risk disease. The study looked at 176 tissues samples covering the spectrum of prostate cancer pathology and compared single candidate biomarkers and derived signatures [Oncotype Dx Genomic Prostate Score (GPS®), Polaris Cell Cycle Score (CCP®) and Decipher®] from low risk tumors with and without the presence of a higher-grade tumor in the prostate. The study concluded that these signatures were not helpful in informing us about the presence of sampled or un-sampled high-grade cancer within the prostate gland, challenging the robustness of these commercially available prognostic markers (43). As such, it is becoming apparent that what you sample is just as important for genomic risk assessment as it is for pathology. This point is emphasized well in a small study of 11 men who were diagnosed with high risk prostate cancer on MRI-US fusion biopsy and underwent radical prostatectomy. Samples were taken from the targeted and random biopsy cores in addition to the radical prostatectomy specimen and derived genomic signatures were compared across the cores (44). The authors reported that the genomics from the targeted biopsy of high Prostate Imaging Reporting & Data System (PIRADS) regions of interest were more in keeping with the genomics from the radical prostatectomy specimen, while the genomics from low PIRADS regions of interest were more in keeping with adjacent benign tissue (45). These results provide further evidence these markers are not immune to the issues of tumor heterogeneity and multi-focality that plaque decision-making using prostate biopsy samples, and mpMRI may play a role in selecting optimal locations for tumor sampling to provide the most prognostic histopathology and genomics. However, we believe the “glass is half full”. While these tests may not overcome the issue of heterogeneity, they do address it and provide a level of risk stratification beyond pathology alone that can be helpful in select cases (46).
Applications to AS
It is well established in the era of mpMRI and MRI targeted biopsy that use of the mpMRI to identify and sample from the region of the prostate that appears most suspicious is likely to yield higher grade cancer compared to random biopsy (47). Furthermore, we are seeing more evidence that the same may be true with respect to genomic risk stratification. It’s been suggested that the largest or most aggressive appearing lesion on mpMRI, or the “index” lesion, may be the tumor most responsible for cancer progression (48). Therefore, given that the pathology from the index lesion best approximates the final pathology from the radical prostatectomy specimen, it may hold true that assessing the genomics from the index lesion may provide the most reliable estimate of the true biology of the cancer. As such, we propose an approach of using mpMRI to evaluate the entire prostate and identify regions or “habitats” of the gland that are suspicious for aggressive prostate cancer so they can be targeted for biopsy to provide pathology and genomics from the tumor most likely to drive progression (Figure 1). We believe this concept will enhance risk assessment and improve outcomes on AS and we are investigating this in an NCI funded, single-institution, interventional prospective trial known as the Miami AS trial (MAST).

Miami AS trial (the MAST trial)
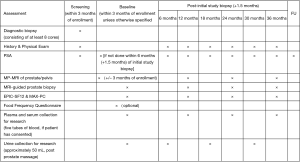
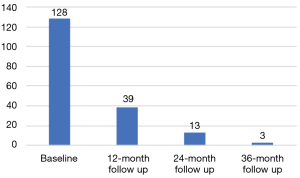
The MAST trial is a single institution, interventional, prospective study evaluating the role of mpMRI and genomic signatures in men with low to intermediate risk prostate cancer who have chosen AS for management of their cancer. The trial enrolls men who were newly diagnosed within the year and have four or less cores of cancer on a minimum 10 core biopsy. Two of the four cores, can be Gleason score 3+4, and there are no exclusions based on the volume of cancer in any core. Gleason score 4+3 in any core, ECE on digital rectal examination (DRE) or inability to obtain mpMRI are exclusion factors for the trial. Men who enroll in the study will have an mpMRI and confirmatory biopsy within 1 year of diagnosis, and every year thereafter for 3 years. If a suspicious lesion (PIRADS 3 or higher) is seen on mpMRI, then a fusion targeted biopsy is performed with two cores from each lesion, in addition to an extended 12-core template biopsy. In men with a negative mpMRI (PIRADS 1-2), only an extended 12-core template is performed. Pathology from both targeted and random cores are selected for whole genome sequencing to assess individual gene expression levels and their derived signatures. Blood for 4K score assessment and post DRE urine is collected annually (Figure 2). To date, we have enrolled 183 patients, of which 128 have undergone their confirmatory baseline biopsy, and 39, 13, and 3 have undergone their 12-, 24-, and 36-month biopsies, respectively (Figure 3). So far, 38 men have progressed on the trial. While the trial is still ongoing, it has already yielded very interesting data, and we expect it to shed some light on the complementary role of mpMRI and genomics in prostate cancer risk stratification. Furthermore, we will also have an opportunity to evaluate various molecular biomarkers to assess their additional role to the clinical armamentarium. Finally, to our knowledge this trial will also be the first to provide prospective data on tumor and genomic heterogeneity pertaining to targeted and random biopsy cores in men undergoing AS for prostate cancer. As a result, we anxiously anticipate the results of this trial and the many question it may help address.
Radiogenomics in risk stratification
The integration of quantitative imaging data (radiomics) to detect correlations with genomic signatures is commonly known as radiogenomics (49). The underlying hypothesis is that mpMRI radiomics features can be used to derive “radiophenotypes” that both correlate to and complement existing validated clinical and genomic risk stratification biomarkers. This concept was introduced by Diehn et al. and Segal et al., by associating extractable features from MRI or CT to global gene expression patterns in glioblastoma multiforme and hepatocellular carcinoma (50,51).
The key in radiogenomic analysis is to be able to connect the gene expression of the prostate tissue with the radiomics features from the location of the tissue. The co-registration of the two types of features is of paramount importance as prostate tumors are heavily heterogeneous (52).
Our team investigated the association of mpMRI radiomics with prostate cancer risk gene expression profiles in mpMRI-guided biopsies tissues (53). Seventeen mpMRI-guided targeted biopsies from six patients were analyzed. The region of interest (ROIs) were identified retrospectively by reevaluating the needle paths of the MRI fusion-guided biopsies. Forty-nine different quantitative features extracted from 3D ROIs based on tumor volumes, intensity, perfusion and diffusion were correlated with genomic profiles associated with poor outcome. We also included radiomics features from the normal appearing tissue in the peripheral zone and transition zone. The radiomics features were associated with the expression of genes on the three testing kits (Oncotype Dx®, Polaris® and Decipher®). There were 445 significant correlations without adjusted p-values but even after adjustment for multiple testing, 64 correlations remained significant (P<0.05). This analysis, albeit the small patient sample, indicates the presence of a strong radiomics association with adverse outcomes. This method could potentially spare invasive diagnostic procedures in the sense of a non-invasive biopsy.
Conclusions
Prostate cancer risk stratification has always relied heavily on clinical factors. The decision to observe or treat prostate cancer has historically been based on the grade and extent of cancer found on biopsy. Recently, we have seen the emergence of mpMRI and various genomic signatures, which have evidence supporting their individual roles in prostate cancer risk stratification, and the selection of patients for AS over immediate treatment. However, we feel the strongest benefit from these tests may come from using them together and we are investigating this currently in a prospective NCI funded clinical trial. We anticipate this research will address many unanswered questions while providing an excellent platform to advance the blossoming field of radiogenomics.
Acknowledgements
Funding: This work was supported in part by National Cancer Institute (R01CA189295 and R01CA190105) and the Stanley J. Glaser Award.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 2009;360:1320-8. [Crossref] [PubMed]
- Hugosson J, Carlsson S, Aus G, et al. Mortality results from the Göteborg randomised population-based prostate-cancer screening trial. Lancet Oncol 2010;11:725-32. [Crossref] [PubMed]
- Dall'Era MA, Albertsen PC, Bangma C, et al. Active surveillance for prostate cancer: a systematic review of the literature. Eur Urol 2012;62:976-83. [Crossref] [PubMed]
- Ahmed HU, El-Shater Bosaily A, Brown LC, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet 2017;389:815-22. [Crossref] [PubMed]
- Fütterer JJ, Briganti A, De Visschere P, et al. Can Clinically Significant Prostate Cancer Be Detected with Multiparametric Magnetic Resonance Imaging? A Systematic Review of the Literature. Eur Urol 2015;68:1045-53. [Crossref] [PubMed]
- Moldovan PC, Van den Broeck T, Sylvester R, et al. What Is the Negative Predictive Value of Multiparametric Magnetic Resonance Imaging in Excluding Prostate Cancer at Biopsy? A Systematic Review and Meta-analysis from the European Association of Urology Prostate Cancer Guidelines Panel. Eur Urol 2017;72:250-66. [Crossref] [PubMed]
- Velasquez MC, Prakash NS, Venkatramani V, et al. Imaging for the selection and monitoring of men on active surveillance for prostate cancer. Transl Androl Urol 2018;7:228-35. [Crossref] [PubMed]
- Clinton TN, Bagrodia A, Lotan Y, et al. Tissue-based biomarkers in prostate cancer. Expert Rev Precis Med Drug Dev 2017;2:249-60. [Crossref] [PubMed]
- Stoyanova R, Takhar M, Tschudi Y, et al. Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res 2016;5:432-47. [Crossref] [PubMed]
- Wilt TJ, Jones KM, Barry MJ, et al. Follow-up of Prostatectomy versus Observation for Early Prostate Cancer. N Engl J Med 2017;377:132-42. [Crossref] [PubMed]
- Hamdy FC, Donovan JL, Lane JA, et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N Engl J Med 2016;375:1415-24. [Crossref] [PubMed]
- Ingimarsson JP, Celaya MO, Laviolette M, et al. Trends in initial management of prostate cancer in New Hampshire. Cancer Causes Control 2015;26:923-9. [Crossref] [PubMed]
- Womble PR, Montie JE, Ye Z, et al. Contemporary Use of Initial Active Surveillance Among Men in Michigan with Low-risk Prostate Cancer. Eur Urol 2015;67:44-50. [Crossref] [PubMed]
- Loeb S, Folkvaljon Y, Curnyn C, et al. Uptake of Active Surveillance for Very-Low-Risk Prostate Cancer in Sweden. JAMA Oncol 2017;3:1393-8. [Crossref] [PubMed]
- Weerakoon M, Papa N, Lawrentschuk N, et al. The current use of active surveillance in an Australian cohort of men: a pattern of care analysis from the Victorian Prostate Cancer Registry. BJU Int 2015;115:50-6. [Crossref] [PubMed]
- Cooperberg MR, Carroll PR. Trends in Management for Patients With Localized Prostate Cancer, 1990-2013. JAMA 2015;314:80-2. [Crossref] [PubMed]
- Prostate cancer, Version 2.2018, NCCN Clinical Practice Guidelines in Oncology. [Internet] 2nd ed. National Comprehensive Cancer Network; 2018 Mar pp. 1-86. Available online: https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- Graham J, Kirkbride P, Cann K, et al. Prostate cancer: summary of updated NICE guidance. BMJ 2014;348:f7524. [Crossref] [PubMed]
- Cooperberg MR, Cowan JE, Hilton JF, et al. Outcomes of active surveillance for men with intermediate-risk prostate cancer. J Clin Oncol 2011;29:228-34. [Crossref] [PubMed]
- Yamamoto T, Musunuru B, Vesprini D, et al. Metastatic Prostate Cancer in Men Initially Treated with Active Surveillance. J Urol 2016;195:1409-14. [Crossref] [PubMed]
- Patel HD, Tosoian JJ, Carter HB, et al. Adverse Pathologic Findings for Men Electing Immediate Radical Prostatectomy. JAMA Oncol 2018;4:89-92. [Crossref] [PubMed]
- Weiner AB, Patel SG, Eggener SE. Pathologic outcomes for low-risk prostate cancer after delayed radical prostatectomy in the United States. Urol Oncol 2015;33:164.e11-7. [Crossref] [PubMed]
- Guo R, Cai L, Fan Y, et al. Magnetic resonance imaging on disease reclassification among active surveillance candidates with low-risk prostate cancer: a diagnostic meta-analysis. Prostate Cancer Prostatic Dis 2015;18:221-8. [Crossref] [PubMed]
- Siddiqui MM, Rais-Bahrami S, Turkbey B, et al. Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 2015;313:390-7. [Crossref] [PubMed]
- Recabal P, Assel M, Sjoberg DD, et al. The Efficacy of Multiparametric Magnetic Resonance Imaging and Magnetic Resonance Imaging Targeted Biopsy in Risk Classification for Patients with Prostate Cancer on Active Surveillance. J Urol 2016;196:374-81. [Crossref] [PubMed]
- Ouzzane A, Renard-Penna R, Marliere F, et al. Magnetic Resonance Imaging Targeted Biopsy Improves Selection of Patients Considered for Active Surveillance for Clinically Low Risk Prostate Cancer Based on Systematic Biopsies. J Urol 2015;194:350-6. [Crossref] [PubMed]
- Stamatakis L, Siddiqui MM, Nix JW, et al. Accuracy of multiparametric magnetic resonance imaging in confirming eligibility for active surveillance for men with prostate cancer. Cancer 2013;119:3359-66. [Crossref] [PubMed]
- Nahar B, Katims A, Barboza MP, et al. Reclassification Rates of Patients Eligible for Active Surveillance After the Addition of Magnetic Resonance Imaging-Ultrasound Fusion Biopsy: An Analysis of 7 Widely Used Eligibility Criteria. Urology 2017;110:134-9. [Crossref] [PubMed]
- Felker ER, Wu J, Natarajan S, et al. Serial Magnetic Resonance Imaging in Active Surveillance of Prostate Cancer: Incremental Value. J Urol 2016;195:1421-7. [Crossref] [PubMed]
- Walton Diaz A, Shakir NA, George AK, et al. Use of serial multiparametric magnetic resonance imaging in the management of patients with prostate cancer on active surveillance. Urol Oncol 2015;33:202.e1-7. [Crossref] [PubMed]
- Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol 2014;66:550-60. [Crossref] [PubMed]
- Cuzick J, Berney DM, Fisher G, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br. J. Cancer 2012;106:1095-9. [Crossref] [PubMed]
- Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12:245-55. [Crossref] [PubMed]
- Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol 2013;31:1428-34. [Crossref] [PubMed]
- Klein EA, Yousefi K, Haddad Z, et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur Urol 2015;67:778-86. [Crossref] [PubMed]
- Cooperberg MR, Davicioni E, Crisan A, et al. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur Urol 2015;67:326-33. [Crossref] [PubMed]
- Spratt DE, Zhang J, Santiago-Jiménez M, et al. Development and Validation of a Novel Integrated Clinical-Genomic Risk Group Classification for Localized Prostate Cancer. J Clin Oncol 2018;36:581-90. [Crossref] [PubMed]
- Klein EA, Haddad Z, Yousefi K, et al. Decipher Genomic Classifier Measured on Prostate Biopsy Predicts Metastasis Risk. Urology 2016;90:148-52. [Crossref] [PubMed]
- Shipitsin M, Small C, Choudhury S, et al. Identification of proteomic biomarkers predicting prostate cancer aggressiveness and lethality despite biopsy-sampling error. Br. J. Cancer 2014;111:1201-12. [Crossref] [PubMed]
- Blume-Jensen P, Berman DM, Rimm DL, et al. Development and clinical validation of an in situ biopsy-based multimarker assay for risk stratification in prostate cancer. Clin Cancer Res 2015;21:2591-600. [Crossref] [PubMed]
- Albala D, Kemeter MJ, Febbo PG, et al. Health Economic Impact and Prospective Clinical Utility of Oncotype DX® Genomic Prostate Score. Rev Urol 2016;18:123-32. [PubMed]
- Wei L, Wang J, Lampert E, et al. Intratumoral and Intertumoral Genomic Heterogeneity of Multifocal Localized Prostate Cancer Impacts Molecular Classifications and Genomic Prognosticators. Eur Urol 2017;71:183-92. [Crossref] [PubMed]
- Hovelson D, Salami SS, Kaplan JB, et al. Integrative molecular profiling challenges robustness of prognostic signature scores in multifocal prostate cancer. J Clin Oncol 2018;36:96. [Crossref]
- Radtke JP, Takhar M, Bonekamp D, et al. Transcriptome Wide Analysis of Magnetic Resonance Imaging-targeted Biopsy and Matching Surgical Specimens from High-risk Prostate Cancer Patients Treated with Radical Prostatectomy: The Target Must Be Hit. Eur Urol Focus 2017. [Crossref] [PubMed]
- Barrett T, Turkbey B, Choyke PL. PI-RADS version 2: what you need to know. Clin Radiol 2015;70:1165-76. [Crossref] [PubMed]
- Dulaney CR, Rais-Bahrami S, Manna DD, et al. DNA repair deregulation in discrete prostate cancer lesions identified on multi-parametric MRI and targeted by MRI/ultrasound fusion-guided biopsy. Oncotarget 2017;8:68038-46. [Crossref] [PubMed]
- Moore CM, Robertson NL, Arsanious N, et al. Image-guided prostate biopsy using magnetic resonance imaging-derived targets: a systematic review. Eur Urol 2013;63:125-40. [Crossref] [PubMed]
- Baco E, Ukimura O, Rud E, et al. Magnetic resonance imaging-transectal ultrasound image-fusion biopsies accurately characterize the index tumor: correlation with step-sectioned radical prostatectomy specimens in 135 patients. Eur Urol 2015;67:787-94. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci USA 2008;105:5213-8. [Crossref] [PubMed]
- Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007;25:675-80. [Crossref] [PubMed]
- Boutros PC, Fraser M, Harding NJ, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 2015;47:736-45. [Crossref] [PubMed]
- Stoyanova R, Pollack A, Takhar M, et al. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies. Oncotarget 2016;7:53362-76. [Crossref] [PubMed]