Genomic biomarkers in prostate cancer
Introduction
Prostate cancer (PCa) is the most common non-cutaneous cancer among men in the United States. In 2018, it is estimated that more than 169,000 men will be diagnosed with prostate cancer and there will be more than 29,000 deaths from the disease in the United States (1). While there is considerable morbidity and mortality as a result of prostate cancer, a significant population of men with prostate cancer have indolent and slow growing disease that does not require intervention. Distinguishing between patients who would benefit from active surveillance (AS), those who require definitive primary treatment, and those who require treatment escalation is a challenging and evolving issue.
While clinically-based nomograms and algorithms have greatly improved risk stratification in PCa, over the last decade there has been a rapid expansion in the field of prognostic and predictive biomarker assays. There are now several FDA approved biomarkers and commercially available clinical laboratory improvement amendments-based (CLIA) tests to help guide clinicians and patients in deciding whether or not biopsy screening is necessary, patients on active surveillance require repeat biopsy, patients require radical prostatectomy (RP) or radiation therapy (RT) as primary treatment, or prognosis for patients who have undergone RP or RT with or without androgen deprivation therapy (ADT).
In this review, we aim to give an update on the current status of liquid and tissue-based FDA-approved and CLIA-based tests available in the United States, the validation studies that led to their development, as well as current shortcomings and gaps in knowledge that exist in the field.
Non-invasive liquid biomarkers
Biomarker tests which sensitively and specifically detect clinically significant prostate cancer from blood or urine samples continue to be pursued. Compared to tissue-based assays liquid biomarkers have minimal risks, as transrectal prostate biopsy carries with it the potential risk of infection and sepsis (2-4). Therefore, the development of novel, non-invasive assays is of great value and importance in the diagnosis and management of PCa. In the following section, we will outline the status of non-invasive urine and serum-based biomarkers which have been develop towards this effort (Figure 1).
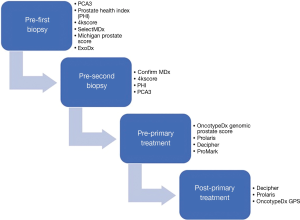
Prostate specific antigen (PSA)-related liquid biomarkers
PSA
Prostate specific antigen is the most commonly used biomarker for the management of PCa today. In the 1980’s PSA was introduced as a biomarker of prostate tissue. Due to the higher sensitivity of PSA relative to prostatic acid phosphatase (PAP) for the detection of PCa, the FDA approved the use of PSA for PCa screening in 1994. Upon its adoption, the upper limit of normal PSA serum level was considered 4.0 ng/mL, although it has since been discovered that approximately 20% of cancers are in patients with PSA levels <4.0 ng/mL (5,6).
PSA is a secreted protein expressed from the gene kallikrein 3. It exists in several isoforms in blood, urine, and semen, all of which are detected by the FDA-approved PSA test. It is expressed in both benign and malignant tissue, and there are several causes for an elevated serum PSA that are not associated with PCa risk (7-9). These include inflammation, trauma, and benign prostate hyperplasia (BPH). The result of common alternative causes for increased PSA reduce the specificity a PCa diagnostic tool. Moreover, at 4 ng/mL, the sensitivity and specificity of PSA are estimated at 67.5–75.0% and 60.3–70.6% respectively (10,11). Although there is much controversy surrounding screening practices with PSA, since its widespread adoption, mortality from prostate cancer has decreased considerably (1). However, the low specificity has also resulted in over-diagnosis and over-treatment of men with indolent prostate cancers that are unlikely to cause morbidity and mortality.
Metrics related to PSA have also been used to improve the sensitivity and specificity of PSA as a biomarker for PCa. Increased PSA density (serum PSA/prostate volume) has been shown to be associated with increased risk of PCa compared to BPH, and risk of higher pathological stage and cancer agressiveness (12,13). Shorter PSA doubling time and PSA velocity have been shown to be associated with increased risk of prostate cancer specific mortality and more aggressive disease course (14,15). PSA stability after RP has also been shown to be associated with biochemical recurrence following RP (16).
The compositions of PSA isoforms are also associated with PCa risk. PSA is normally complexed with the protein antichymotrypsin (ACT) in the blood, with a fraction of PSA being uncomplexed in the serum (fPSA) (17). In PCa, the percent of fPSA is lower than in patients without PCa. %fPSA is FDA approved for use in patients with a PSA of 4–10 ng/mL to increase the sensitivity and specificity of PSA testing (17). It also increases the sensitivity and specificity in lower ranges of PSA (2–4 ng/mL) (18). There are drawbacks to fPSA however. It requires processing within 24 hours to yield reliable results, and procedures such as prostate biopsy or DRE can affect the levels of serum fPSA (19,20). Thus, it has been difficult for clinicians to adopt and implement fPSA as a screening test in urologic practices.
Since the widespread adoption of PSA as a screening and monitoring test, a great deal of effort has been made to improve the sensitivity and specificity of the serum-based marker. The Prostate Health Index and 4kscore utilize isoforms of PSA and its precursors to help risk-stratify patients with an abnormal PSA.
Prostate Health Index (Beckman Coulter, Brea, CA)
Prostate Health Index (PHI) is a mathematical formula developed by Beckman Coulter (Brea, CA) that was designed to better identify patients with a slightly elevated PSA (2.0–10.0 ng/mL) who are at high risk of harboring prostate cancer. The formula incorporates both serum [−2]proPSA, which is the primary precursor PSA isoform prevalent in malignant tissue than benign prostate tissue, and serum free PSA which has been shown to be more specific that total tPSA in the detection of PCa (17,21). PHI is calculated with the formula
4Kscore test (Opko Lab, Miami, FL)
4Kscore test is an algorithm that combines serum concentrations of a “4 kallikrein (4k) panel” [total PSA (tPSA), free PSA (fPSA), intact PSA (iPSA), and human kallikrein 2 (hK2)], with clinical information (age, DRE status, previous biopsy result) to predict the likelihood of detecting high-grade PCa (GS ≥7) on prostate biopsy. The test was first developed in Europe as part of the European Randomized Study of Prostate Cancer Screening (ERSPC), where the measurement of tPSA, fPSA, iPSA and hK2 were consistently correlated with positive biopsy in separate cohorts (25-28). In a cohort of 740 biopsy-naïve with a PSA ≥3.0 ng/mL, adding the 4k panel to clinical variables increased the AUC from 0.87 (95% CI: 0.78–0.92) to 0.90 (95% CI: 0.86–0.96) with the primary outcome of high-grade cancer (29). In a similar group of 262 men, including the 4k panel increased the AUC from 0.77 (95% CI: 0.69–0.85) to 0.87 (95% CI: 0.81–0.93) compared to clinical variables alone (27).
The 4K panel has also been evaluated in men with a history of a negative biopsy. In a cohort of 925 men with elevated PSA, the addition of the 4k panel increased the AUC from 0.76 to 0.87 compared to clinical factors alone (26). In this cohort, at a cutoff of 15% risk, 712 repeat biopsies could have been avoided while delaying the diagnosis of 3 cancers of GS 7 or greater.
Of note, in 2015 a study was performed by Nördstrom et al. which compared the diagnostic performance of the 4k panel, PHI, and a base model of PSA and age. The trial included 531 biopsy naïve men with PSA levels between 3 and 15 ng/mL. The trial showed that 4k and PHI were comparable, with AUC’s of 0.718 and 0.711 respectively for predicting GS ≥7 cancer. Both algorithms performed better than age and PSA for the prediction of high-grade cancer.
OPKO Health commercialized the 4k panel in the US, and sponsored a validation study that included 1,012 men undergoing prostate biopsy at 26 independent sites (30-32). 4Kscore is identical to the 4 kallikrein assays investigated in Europe, aside from the inclusion of biopsy history as well as different sources for the assays of tPSA, fPSA, iPSA, and hK2. The AUC of 4Kscore in the prediction of GS ≥7 was 0.818 (95% CI: 0.788–0.849) which was slightly improved by the addition of a DRE (AUC =0.821; CI 0.794–0.852) (30,32). In the cohort of 1,012, using a 4Kscore cutoff of 7.5% risk, 360 biopsies could have been avoided while missing 16 of 215 aggressive cancers found on biopsy (30). 4Kscore was also associated with higher grade in patients from the same cohort who underwent RP (n=144). However, this association was not significant when adjusting for D’Amico risk or Cancer of the Prostate Risk Assessment (CAPRA) (31).
The 4k panel has also been studied in the setting of active surveillance. As part of the Canary PASS trial, Lin et al. carried out using the 4k panel to analyze the serum of men on active surveillance. The setting in this study is unique from past analyses of the 4k panel, in that the assay was used on men already diagnosed with PCa. As a predictor for reclassification on active surveillance (GS 6 to GS ≥7 on a subsequent biopsy), the 4k panel did not perform markedly better than PSA when combined with a clinical model consisting of age, body mass index, history of negative biopsies, percentage of positive biopsy cores, and prostate volume (33). At the time of first surveillance biopsy, the 4k + clinical model had an AUC of 0.783, while the PSA + clinical model had an AUC of 0.740. At the time of subsequent biopsies, there was no difference in the AUC of the two models. So, while the 4k panel is more accurate than clinical factors in the pre-diagnosis setting, there is minimal benefit of 4k once clinical tumor parameters are known.
Non-PSA liquid biomarkers
A number of non-PSA liquid biomarkers have been developed with the goal of improving diagnostic accuracy in men with suspicion of prostate cancer. They are detailed in the following section.
PCA3 (Hologic, Marlborough, MA)
The prostate cancer antigen 3 (PCA3) assay from Progensa is a urine-based, post-DRE, FDA approved test designed to assess the need to re-biopsy men with a previous negative biopsy. PCA3 is a long noncoding RNA that is specific to malignant prostate tissue, and is overexpressed in 95% of prostate cancers with a 66-fold median up-regulation (29,34,35). Unlike PSA, PCA3 expression is independent of prostate volume (36). Several studies of men post-DRE and on active surveillance (AS) observed a strong positive correlation between PCA3 levels in the urine and positive biopsy on repeat biopsy (36-38). In the trial for FDA approval, the post DRE urine samples, and serum from 466 men with at least 1 prior negative biopsy were analyzed for PCA3 and PSA RNA levels. On MVA, PCA3 score [(PCA 3 RNA copies per mL/PSA RNA copies per mL) ×1,000] of greater than 25 was associated with increased risk of positive repeat biopsy (OR =4.56; 95% CI: 2.65–7.84; P<0.0001) (36). When combined with clinical factors such as age, DRE result, family history of PCa, race, serum PSA, and number of previous negative biopsies, the AUC of the PCA3 score increased from 0.653 to 0.740 (P=0.0007) compared to the clinical factors alone.
In reality, PCA3 did not vastly outperform PSA based on a dichotomous cutoff of 25 as proposed in the FDA validation trial (36,38,39). However, in a study published by Wei et al. PCA3 demonstrated a negative predictive value (NPV) for any prostate cancer of 88% on repeat biopsy with a cutoff of PCA<20, and a positive predictive value (PPV) of 80% on initial biopsy with a cutoff of PCA3>60 (40). So, while there may not be tremendous value with a single dichotomous cut-off, PCA3 may provide valuable information when its value is either very high or very low.
Mi-Prostate Score (MLabs, Ann Arbor, MI)
MiPS is an algorithm developed at the University of Michigan that combines urine TMPRSS2:ERG, prostate cancer antigen 3 (PCA3), and serum PSA to predict the risk of detecting PCa on biopsy. The algorithm uses an FDA approved assay in the Progensa PCA3 test (Hologic Inc, Bedford, MA, USA), and a clinical-grade assay for TMPRSS2:ERG (41). TMPRSS2:ERG is a gene fusion that is present approximately 50% of PCa patients. The role of TMPRSS2:ERG in the development of PCa is not known (17). PCA3 is a long noncoding RNA that is overexpressed in 95% of prostate cancers. The median up-regulation of PCA3 in PCa is 66-fold (29,35). In the initial study demonstrating the utility of PCA3, TMPRSS2:ERG, and PSA as combined biomarkers of PCa, serum and post-DRE urine samples were collected on 48 men undergoing prostate biopsy. The MiPS multivariable algorithm was more specific than any of the individual variables, with an AUC of 0.88 and a specificity of 90% and an 80% sensitivity (41). The MiPS algorithm was validated in larger cohort (n =1,225) and demonstrated an AUC =0.751 detecting PCa on biopsy, and an AUC =0.772 detecting GS 7 or higher. This was significantly better than PSA alone (AUC =0.651 for GS 7 for high grade cancer). Of note, the validation of MiPS did not compare its diagnostic accuracy to that of PCA3 alone, which is has already been shown to outperform PSA. So while MiPS certainly provides value over PSA alone, its value over PCA3 still requires validation.
Select MDx (MDx Health, Irvine, CA)
Select MDx is an algorithm that combines clinical factors with a urine-based assay for RNA levels of HOXC6 and DLX1. In the trial that validated the assay to detect clinically significant PCa, 519 patients undergoing biopsy were prospectively enrolled as the training cohort. A post-DRE urine sample was analyzed for the expression of five known markers of PCa and KLK3 (as a control RNA) (42). Of the five markers, combining HOXC6 and DLX1 resulted in the highest AUC (0.76; 95% CI: 0.71–0.81) with the primary outcome of GS ≥7 on biopsy. Of note, PCA3 (AUC =0.68) was also included in the analysis. However, HOXC6 and DLX1 more reliably predicted clinically significant PCa. The Select MDx metric was validated in a separate cohort (n=386) in which the combination of HOXC6 and DLX1 had an AUC of 0.73 (95% CI: 0.67–0.78). In the final model that included PSAD, DRE, PSA, age, history of biopsy, and family history of PCa, the AUC was 0.90 (0.87–0.93) for high-grade PCa. In the validation cohort the NPV was 94%, confirming the clinical utility of the biomarkers in reliably predicting patients that are unlikely to have clinically significant PCa (43). It should be noted that markers such as Select MDx and 4kscore provide an advantage over markers such as PHI, in that they incorporate clinical variables into their algorithm to be used as a single metric.
ExoDx Prostate Intelliscore (Exosome Diagnostics, Waltham, MA)
ExoDx Prostate Intelliscore (EPI) is a pre-biopsy RNA-based assay which uses the expression of PCA3 and ERG isolated from urinary exosomes as a predictor of men who will have GS 7 or greater PCa on diagnostic biopsy. Exosomes are vesicles secreted by cells, and contain proteins, RNA, and other molecules which are closely representative of the content of cellular origin (44). PCa cells secrete more exosomes than non-cancerous cells of the prostate, and in cell-lines from other cancer types, it has been shown that the RNA which is secreted in exosomes can be upregulated several hundred-fold when compared to the cell of origin (45). It has been previously shown that post-DRE urine samples contain exosomes with PCA3 and TMPRSS2: ERG mRNA (44). However, EPI does not require a DRE to isolate the exosomal RNA, which is a potential benefit to patients undergoing testing. EPI was validated in a training cohort of 255 men and a validation cohort of 519 men who met the following criteria: age ≥50, PSA 2–10 ng/mL, and never having undergone prostate biopsy prior to study participation (46). They looked at the performance of the assay to predict GS ≥7 on prostate biopsy, and found in the validation cohort that when EPI was combined with clinical characteristics of age, PSA, race, and family history of prostate cancer, EPI demonstrated an AUC of 0.73 (95% CI: 0.68–0.77) compared to the AUC of 0.63 (95% CI: 0.58–0.68) of clinical characteristics alone. With the authors’ proposed cutoff, the model yielded a sensitivity, specificity, PPV, and NPV of 91.89, 33.96, 35.70, and 91.30, respectively, in the validation cohort. If used as a determinant of who should and should not get a biopsy, EPI could have reduced the number of biopsies by 27% while missing 5% of dominant Gleason pattern 4 or greater PCa.
Tissue-based biomarkers
While great strides have been made in the development of “liquid” biomarkers, tissue-based assays remain the gold standard for accurately representing the tumor environment in PCa. In the last several years, tissue-based biomarkers have become standard of care for patients undergoing biopsy and radical prostatectomy. Confirm MDx, Decipher, Prolaris, and OncotypeDx GPS are now recommended by the Molecular Diagnostic Services Program which aims to identify and establish coverage and insurance reimbursement for molecular diagnostic tests. In addition, these tests and are now included in the NCCN guidelines for the management of PCa, which is not yet the case for liquid biomarker assays. A challenging aspect of these tests however, is that they are validated in many of the same contexts. Just like in the pre-diagnosis setting, deciding which test a physician should utilize after prostate biopsy or RP remains challenging. In the following section we will review in which contexts these assays were originally designed and validated, as well as other contexts in which they may play a useful role in patient management (Table 1).
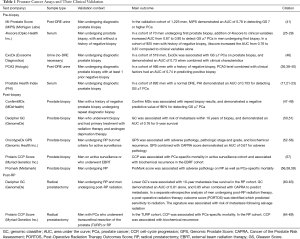
Full table
Confirm MDx (MDx Health, Irvine CA)
Confirm MDx is an epigenetic-based assay that was developed to aid in the management of patients who have a history of a negative biopsy and are considering re-biopsy (47-49). MDx utilizes a modified polymerase chain reaction (multiplex quantitative methylation specific PCR) to determine the DNA methylation status of three known tumor suppressor genes, GSTP1, APC, and RASSF1. The assay is based on the hypothesis that normal tissue adjacent to a tumor may harbor epigenetic, genetic, or protein expression changes, and that monitoring the state of these non-tumor cells can decrease the likelihood of a false-negative result on biopsy (70). The initial study used to develop Confirm MDx analyzed samples from 498 subjects of the MATLOC (Methylation Analysis to Locate Occult Cancer) study who had a negative initial screening biopsy and a repeat biopsy that was either positive or negative. The Confirm MDx assay was associated with a NPV of 90% (95% CI: 87–93%), and in an MVA controlling for age PSA, DRE, and first biopsy result, the epigenetic assay was independently associated with cancer detected on repeat biopsy (OR 3.17; 95% CI: 1.81–5.53) (47). In the DOCUMENT multicenter trial, 350 patients with negative screening biopsy underwent the epigenetic assay. A negative epigenetic assay was associated with a NPV of 88% (95% CI: 85–91%), and an OR of 2.69 (95% CI: 1.60–4.51) in an MVA (48). The cohorts from the MATLOC and the DOCUMENT trials were combined to evaluate the association between DNA-methylation intensity and diagnosis of GS ≥7 on repeat biopsy. Confirm MDx was associated with a NPV of 96%, and in methylation positive men, the degree of methylation was significantly higher in those with GS ≥7 compared to those with GS ≤6 (49).
Prolaris (Myriad Genetics Inc., Salt Lake City, UT)
Prolaris is a gene expression assay that measures the expression of 31 cell cycle progression (CCP) genes and 15 housekeeping genes. The test has been validated in prostate needle biopsies as well as RP samples in its association with 10-year prostate cancer-specific mortality risk (64-69). The initial study compared patients with PCa undergoing transurethral resection of the prostate (TURP) with patients undergoing RP. In the 366 patients who underwent RP, each unit increase in CCP was associated with biochemical recurrence on univariate (HR: 1.89; 95% CI: 1.54–2.31; P<0.0001) and multivariate analysis (HR: 1.77; 95% CI: 1.40–2.22; P<0.0001). In the TURP cohort, each unit increase (equivalent to a doubling of gene expression) in CCP was associated with prostate specific mortality on univariate (HR: 2.92; 95% CI: 2.38–3.57; P<0.0001) and multivariate analysis (HR: 2.56; 95% CI: 1.85–3.53; P<0.001) (64). In a study of 413 post-RP patients, a combination of CAPRA-S and CCP scores was more closely associated with recurrence risk compared to either of the metrics alone (65).
CCP was also validated in biopsy samples in a cohort managed by AS. In this cohort of 349, a one-unit increase in CCP was associated with PCa-specific mortality on univariate (HR: 2.02; 95% CI: 1.62–2.53; P<0.0005) and multivariate analysis (HR: 1.65; 95% CI: 1.31–20.9; P<0.0001) controlling for clinical variables (66). In a cohort of 141 men who underwent external beam radiation therapy as primary curative therapy, CCP score on needle biopsy samples was associated with increased risk of biochemical recurrence on univariate (HR: 2.55; 1.43–4.55; P=0.0017) and multivariate analysis (HR: 2.11; 1.05–4.25; P=0.034) when controlling for clinical variables including combination ADT and EBRT (57).
Decipher RP and Decipher Biopsy (GenomeDx, Vancouver BC, Canada)
Decipher is a 22-gene panel developed by GenomeDx (Vancouver BC, Canada) as a genomic classifier (GC) prediction model for metastasis (60). Transcriptome-wide RNA microarray sequencing including more than 240,000 known protein coding genes and non-protein coding RNAs was performed on 545 RP samples from patients with median follow-up of 16.9 years. Patients were classified into three groups: no biochemical recurrence (BCR) or clinical metastasis within 5 years of RP, BCR within 5 years of RP with no clinical metastasis, or clinical metastasis within 5 years and used in training and validation sets. Logistic regression of the microarray results yielded 43 RNA transcripts that were differentially expressed between cases and controls, of which 22 improved the performance of a random forest machine learning algorithm in the GC. These 22 biomarkers include both protein coding and non-coding RNAs, with roles in cell proliferation, cell-cycle progression, immune response, cell structure, cell adhesion, and motility, as well as three genes with unknown function. The 22-gene model yielded an AUC of 0.90 and 0.75 in the training and validation sets respectively, which was better than any individual clinical or genomic variable on its own (60). The decipher GC has also been validated in a high-risk group of PCa patients (PSA >20 ng, Gleason ≥8, pT3b, or Mayo Clinic nomogram score of 10 or greater), where the AUC of the GC was 0.79 for predicting 5-year metastasis after RP. When stratified into low, intermediate, and high GC scores, the incidence of metastasis in this high-risk group was 2.4%, 6.0%, and 22.5% respectively (P<0.001) (61).
Recently, the first individual patient level meta-analysis has been performed for Decipher of various risk groups of patients undergoing RP. This meta-analysis by Spratt et al. demonstrated that Decipher independently was prognostic for the development of metastasis. This finding was maintained when looking at subgroups by race, clinical or pathologic subgroups, and treatment subgroups using adjuvant or salvage therapies (71).
High decipher GC scores have been validated on biopsy cores as a predictor of metastasis and prostate cancer specific mortality within 10 years of RP or RT. On multivariate analysis (MVA), an increase in biopsy decipher score of 0.1 (scored 0–1.0) was associated with an increased risk of metastasis (HR: 1.72; 95% CI: 1.07–2.81; P=0.02) (50). In 100 patients with intermediate and high-risk PCa who underwent RT and ADT as primary treatment, each 0.1 unit increase in GC score associated with time to distant metastasis on multivariable analysis (HR: 1.36; 95% CI: 1.10–1.84; P=0.006) after adjusting for clinical variables (51). In the MVA, neither CAPRA, NCCN, nor Gleason score were statistically significant in their association with time to metastasis. The survival C-index (5 years post-RT) for Biopsy decipher, CAPRA, and NCCN, were 0.76 (0.57–0.89), 0.45 (0.27–0.64), and 0.63 (0.4–0.78) respectively in this cohort.
The Decipher test has also been used to develop a novel clinical-genomic NCCN risk grouping system (72). Using nearly 7,000 patients, including nearly 1,000 prospective biopsy samples, pre-treatment clinical-genomic risk groups were trained and validated for both metastasis and PCSM. In the validation cohort the AUC to predict metastasis was approximately 0.6–0.7 for NCCN or CAPRA, which increased to 0.84 for the clinical-genomic risk groups.
A benefit of the technology from GenomeDx, is their ability to look at other RNA expression signatures, which may be associated with outcomes beyond the scope of their original gene expression panel. Retrospectively, GenomeDx has used their technology to identify signatures related to other treatment modalities such as radiation, androgen deprivation therapy (ADT), chemotherapy sensitivity (62,63). The Post-Operative Radiation Therapy Outcomes Score (PORTOS) score is a 24-gene panel that was developed to predict response to RT following RP. RT patients were retrospectively matched with patients who had not received radiation, controlling for Gleason score, PSA, concentration, surgical staging, and treatment ADT. In a training cohort of 198 patients, 1,800 DNA damage repair and radiation response genes were ranked using an Affymetrix Human Exon 1.0 ST microarray (Affymetrix, Santa Clara, CA). 24 genes were identified that were predictive of EBT response. In patients with a high PORTOS score, those who underwent RT had decreased incidence of metastasis compared to those who did not (HR: 0.12; 95% CI: 0.03–0.41; P<0.0001). In patients with a low PORTOS score, those who received RT had an increased incidence of metastasis compared to those who did not undergo radiotherapy (HR: 2.6; 95% CI: 1.6–4.1; P<0.0001). A similar trend was seen in the validation cohort where high PORTOS (HR: 0.15; 95% CI: 0.04–0.60; P=0.0020) was associated with decreased incidence of metastasis with radiation. However, in this cohort, the response in the low PORTOS score group (HR: 0.92; 95% CI: 0.56–1.51; P=0.76) was not significant. PORTOS represents the first gene panel of its kind, in that it is a predictive biomarker panel rather than prognostic. PORTOS has yet to be validated prospectively in a randomized control trial, and its association with RT sensitivity has only been shown in post-RP patients. Therefore, more investigation is required to validate the gene panel prospectively, and also in the setting of RT for primary treatment of prostate cancer.
Another prognostic and predictive biomarker that has resulted from the Decipher platform is PAM50 which has identified three primary subtypes of prostate adenocarcinoma. PAM50 was originally developed to subtype breast cancer into luminal and basal subtypes. The same clustering signature was used to identify luminal A, luminal B, and basal subtypes in prostate cancer (62). In a study of 3,728 patients, it was discovered that patients with the luminal B subtype demonstrated the poorest clinical prognosis, with greater risk of biochemical recurrence, prostate cancer specific mortality, development of distant metastases, and worse overall survival. PAM50 subtyping also predicts response to ADT. In a sub-analysis of 315 post-RP patients who were matched for clinical characteristics and compared for response to ADT (210 men who did not receive ADT, and 105 who did), luminal B subtype predicted response to ADT. 33% of men with luminal B cancers who were treated with ADT went on to develop distant metastasis vs. 55% in the no ADT controls (P=0.06) (62). Remarkably, the same subtypes can be identified even among men with histologically homogeneous, low-risk tumors. (Cooperberg et al. Eur Urol in press) Similar to PORTOS, PAM50 has not been validated prospectively for response to ADT based on luminal/basal subtyping. However, it certainly warrants more investigation, as predicting response to ADT would be incredibly valuable for clinicians. The use of PAM50 is being incorporated into the currently open phase 2 randomized NRG GU-006 to prospectively assess the prognostic and predictive capacity of this biomarker for men undergoing post-operative radiotherapy and the benefit of hormone therapy by subtype.
Oncotype Dx (Genomic Health, Redwood City, CA)
Oncotype Dx Genomic Prostate Score (GPS) is based on the levels of expression of 17 genes in tissue obtained by needle biopsy and is designed to help guide treatment decisions in men who are considering active surveillance (AS) vs. primary treatment (52). To identify the seventeen genes used in the assay, the expression levels of 727 genes were analyzed in a group of patients that had undergone RP (n=441). Controlling for GS and AUA risk, expression changes for 198 of the genes correlated with recurrence, PCa death, and adverse pathology at prostatectomy in both unifocal and multifocal disease. For validation in needle biopsy samples, the expression of 81 of the genes associated with aggressive disease from the RP cohort were analyzed in 167 AUA low- and intermediate-risk patients with both biopsy and RP samples. Of the 81 genes, 58 positively correlated with adverse pathology at RP, and consistency between the biopsy and RP samples. From the 58 genes, 12 genes were selected based on their consistency across the various samples, reproducibility based on high expression and dynamic range, and possible relationship to four pathways known to be involved in PCa progression (stromal expression, cellular organization, androgen pathway, and proliferation). These expression patterns are combined with five reference genes in an algorithm to yield a Genomic Prostate Score (GPS) (53).
To assess the utility of GPS as an aid to decision making in patients on AS, a validation study at the University of California San Francisco was performed on 395 patients who underwent RP, but met criteria for AS. In these patients, GPS was a significantly associated with pathologic stage and grade on RP (OR: 2.1; 95% CI: 1.4–3.2; P<0.001). Combining GPS and CAPRA improved the AUC (0.67) for favorable pathology compared to CAPRA alone (52). In a single independent validation cohort, GPS score was determined in the biopsies 431 men. In a multivariate analysis, each 20-unit increase in GPS (scored 0–100) was associated with time to biochemical recurrence (HR: 2.7; P<0.001), and adverse pathology (OR: 3.3; P<0.001) on RP in both Caucasian and African American men (54). Of note, a 20-unit increased in GPS represents a dramatic difference, as the independent validation study showed IQR’s with magnitudes of 15–17 GPS units. As a result, a 20-unit increase may not be the most clinical valuable metric for GPS. In addition, while GPS was validated in patients that were eligible AS who underwent RP, GPS has yet to be validated in an actual AS cohort. However, preliminary analysis would suggest that GPS is associated with biopsy upgrade (GS 6 to GS ≥7 on subsequent biopsy) (72).
Promark (Metamark Genetics, Waltham, MA)
Promark is an automated immunofluorescence-based assay designed to guide treatment decisions in men with GS 3+3 or GS 3+4 cancer (56,58,59). Promark measures the expression of eight proteins (DERL1, CUL2, SMAD4, PDSS2, HSPA9, FUS, phosphorylated S6, and YBOX1) associated with adverse pathology on RP, defined as surgical GS ≥3+4, or nonlocalized disease (> T3a, ≥ N1, or ≥ M1). The design of Promark addresses inconsistency in biopsy samples that is a consequence of the expected variability in sample constitution and pathologist discordance in Gleason scoring (73,74). The Promark analysis identified the areas of lowest and highest grade in the biopsies of 380 patients who had undergone RP and had long-term follow-up data, and quantified the expression of 39 candidate proteins that had known biological relevance for cancer. These 39 proteins were evaluated for antibody specificity, signal intensity, dynamic rage, and stability, and 12 were identified as significantly associated with adverse pathology on RP, and prostate cancer specific mortality in both the low-grade and high-grade biopsy samples on univariate and multivariate analysis. The AUC with the primary outcome of adverse pathology on RP was 0.72 (95% CI: 0.64–0.79) in low-grade biopsies, and 0.70 (95% CI: 0.62–0.77) in the high-grade biopsies (65). From the 12-gene panel, an 8 biomarkers assay was validated in its association with adverse pathology on RP specifically in GS 3+3 and GS 4+4 biopsy samples. The AUC for favorable pathology prediction when combined with NCCN and D’Amico risk stratification were 0.75 (95% CI: 0.69–0.81) and 0.75 (95% CI: 0.69–0.81), respectively.
Conclusions
In this review, we have highlighted the numerous biomarker-based clinical tests which are available at nearly every stage of PCa. From pre-diagnosis, to post-primary treatment, it can be challenging for patients and clinicians alike to navigate the spectrum of tests available. While these tests offer considerable information to guide treatment decisions, they also can be costly and contribute to financial toxicity.
One population in which there are currently no prospectively validated liquid or tissue-based biomarker assays is patients on active surveillance. Once patients enroll on active surveillance, continual surveillance with annual or biannual biopsy is burdensome and costly to payers and patients. It would be incredibly valuable to have validated prognostic biomarkers which might allow for the delay of screening biopsy with a strong NPV, or predict patients that are likely to fail active surveillance with a strong PPV.
To date there are also no head-to-head studies comparing the prognostic capabilities of tests like Decipher and Prolaris, which can be used in the same patient population and provide risk projections of similar outcomes. To determine which biomarker assays are the most effective at each stage of disease progression would ideally require randomized control trials and would take years to decades of follow-up to determine outcomes. Systematic analysis of markers via prospective registries may help demonstrate the extent to which markers do or do not improve decision-making and outcomes. In the coming years, it is important for clinicians to stay aware of the developments of this rapidly-growing field, as choosing the right test for the right patient may become an increasingly nuanced decision.
Acknowledgements
We thank all members of the Feng lab for their contributions.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Borghesi M, Ahmed H, Nam R, et al. Complications After Systematic, Random, and Image-guided Prostate Biopsy. Eur Urol 2017;71:353-65. [Crossref] [PubMed]
- Lundström KJ, Drevin L, Carlsson S, et al. Nationwide Population Based Study of Infections after Transrectal Ultrasound Guided Prostate Biopsy. J Urol 2014;192:1116-22. [Crossref] [PubMed]
- Bruyère F, Malavaud S, Bertrand P, et al. Prosbiotate: A Multicenter, Prospective Analysis of Infectious Complications after Prostate Biopsy. J Urol 2015;193:145-50. [Crossref] [PubMed]
- Catalona WJ, Smith DS, Ornstein DK. Prostate cancer detection in men with serum PSA concentrations of 2.6 to 4.0 ng/mL and benign prostate examination. Enhancement of specificity with free PSA measurements. JAMA 1997;277:1452-5. [Crossref] [PubMed]
- Thompson IM, Pauler DK, Goodman PJ, et al. Prevalence of Prostate Cancer among Men with a Prostate-Specific Antigen Level ≤4.0 ng per Milliliter. N Engl J Med 2004;350:2239-46. [Crossref] [PubMed]
- Lintula S, Stenman J, Bjartell A, et al. Relative concentrations of hK2/PSA mRNA in benign and malignant prostatic tissue. Prostate 2005;63:324-9. [Crossref] [PubMed]
- Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat Rev Cancer 2008;8:268-78. [Crossref] [PubMed]
- Lilja H. Biology of prostate-specific antigen. Urology 2003;62:27-33. [Crossref] [PubMed]
- Brawer M, Lange PH. PSA in the screening, staging and follow-up of early-stage prostate cancer. World J Urol 1989;7:7-11. [Crossref]
- Brawer MK, Benson MC, Bostwick DG, et al. Prostate-specific antigen and other serum markers: current concepts from the World Health Organization Second International Consultation on Prostate Cancer. Semin Urol Oncol 1999;17:206-21. [PubMed]
- Benson MC, Whang IS, Pantuck A, et al. Prostate specific antigen density: a means of distinguishing benign prostatic hypertrophy and prostate cancer. J Urol 1992;147:815-6. [Crossref] [PubMed]
- Kundu SD, Roehl KA, Yu X, et al. Prostate specific antigen density correlates with features of prostate cancer aggressiveness. J Urol 2007;177:505-9. [Crossref] [PubMed]
- Teeter AE, Griffin K, Howard LE, et al. Does Early Prostate Specific Antigen Doubling Time after Radical Prostatectomy, Calculated Prior to Prostate Specific Antigen Recurrence, Correlate with Prostate Cancer Outcomes? A Report from the SEARCH Database Group. J Urol 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Moreira DM, Presti JC Jr, Aronson WJ, et al. Natural history of persistently elevated prostate specific antigen after radical prostatectomy: results from the SEARCH database. J Urol 2009;182:2250-5. [Crossref] [PubMed]
- Koulikov D, Mohler MC, Mehedint DC, et al. Low detectable prostate specific antigen after radical prostatectomy--treat or watch? J Urol 2014;192:1390-6. [Crossref] [PubMed]
- Catalona WJ, Partin AW, Slawin KM, et al. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: a prospective multicenter clinical trial. JAMA 1998;279:1542-7. [Crossref] [PubMed]
- Haese A, Dworschack RT, Partin AW. Percent free prostate specific antigen in the total prostate specific antigen 2 to 4 ng/ml. range does not substantially increase the number of biopsies needed to detect clinically significant prostate cancer compared to the 4 to 10 ng/ml. range. J Urol 2002;168:504-8. [Crossref] [PubMed]
- Woodrum D, French C, Blair Shamel L. Stability of free prostate-specific antigen in serum samples under a variety of sample collection and sample storage conditions. Urology 1996;48:33-9. [Crossref] [PubMed]
- Ornstein DK, Rao GS, Smith DS, et al. Effect of Digital Rectal Examination and Needle Biopsy on Serum Total and Percentage of Free Prostate Specific Antigen Levels. J Urol 1997;157:195-8. [Crossref] [PubMed]
- Mikolajczyk SD, Marker KM, Millar LS, et al. A truncated precursor form of prostate-specific antigen is a more specific serum marker of prostate cancer. Cancer Res 2001;61:6958-63. [PubMed]
- Catalona WJ, Partin AW, Sanda MG, et al. A multicenter study of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ml prostate specific antigen range. J Urol 2011;185:1650-5. [Crossref] [PubMed]
- de la Calle C, Patil D, Wei JT, et al. Multicenter Evaluation of the Prostate Health Index to Detect Aggressive Prostate Cancer in Biopsy Naive Men. J Urol 2015;194:65-72. [Crossref] [PubMed]
- Lazzeri M, Abrate A, Lughezzani G, et al. Relationship of chronic histologic prostatic inflammation in biopsy specimens with serum isoform [-2]proPSA (p2PSA), %p2PSA, and prostate health index in men with a total prostate-specific antigen of 4-10 ng/ml and normal digital rectal examination. Urology 2014;83:606-12. [Crossref] [PubMed]
- Vickers AJ, Cronin AM, Aus G, et al. Impact of recent screening on predicting the outcome of prostate cancer biopsy in men with elevated prostate-specific antigen: data from the European Randomized Study of Prostate Cancer Screening in Gothenburg, Sweden. Cancer 2010;116:2612-20. [PubMed]
- Gupta A, Roobol MJ, Savage CJ, et al. A four-kallikrein panel for the prediction of repeat prostate biopsy: data from the European Randomized Study of Prostate Cancer screening in Rotterdam, Netherlands. Br J Cancer 2010;103:708-14. [Crossref] [PubMed]
- Benchikh A, Savage C, Cronin A, et al. A panel of kallikrein markers can predict outcome of prostate biopsy following clinical work-up: an independent validation study from the European Randomized Study of Prostate Cancer screening, France. BMC Cancer 2010;10:635. [Crossref] [PubMed]
- Vickers AJ, Cronin AM, Aus G, et al. A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: data from the European Randomized Study of Prostate Cancer Screening in Goteborg, Sweden. BMC Med 2008;6:19. [Crossref] [PubMed]
- Salagierski M, Schalken JA. Molecular diagnosis of prostate cancer: PCA3 and TMPRSS2:ERG gene fusion. J Urol 2012;187:795-801. [Crossref] [PubMed]
- Zappala SM, Dong Y, Linder V, et al. The 4Kscore blood test accurately identifies men with aggressive prostate cancer prior to prostate biopsy with or without DRE information. Int J Clin Pract 2017.71. [PubMed]
- Punnen S, Nahar B, Prakash NS, et al. The 4Kscore Predicts the Grade and Stage of Prostate Cancer in the Radical Prostatectomy Specimen: Results from a Multi-institutional Prospective Trial. Eur Urol Focus 2017;3:94-9. [Crossref] [PubMed]
- Parekh DJ, Punnen S, Sjoberg DD, et al. A multi-institutional prospective trial in the USA confirms that the 4Kscore accurately identifies men with high-grade prostate cancer. Eur Urol 2015;68:464-70. [Crossref] [PubMed]
- Lin DW, Newcomb LF, Brown MD, et al. Evaluating the Four Kallikrein Panel of the 4Kscore for Prediction of High-grade Prostate Cancer in Men in the Canary Prostate Active Surveillance Study. Eur Urol 2017;72:448-54. [Crossref] [PubMed]
- Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999;59:5975-9. [PubMed]
- Hessels D, Klein Gunnewiek JM, van Oort I, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol 2003;44:8-15; discussion 15-6. [Crossref] [PubMed]
- Haese A, de la Taille A, van Poppel H, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol 2008;54:1081-8. [Crossref] [PubMed]
- Wu AK, Reese AC, Cooperberg MR, et al. Utility of PCA3 in patients undergoing repeat biopsy for prostate cancer. Prostate Cancer Prostatic Dis 2012;15:100-5. [Crossref] [PubMed]
- Marks LS, Fradet Y, Deras IL, et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology 2007;69:532-5. [Crossref] [PubMed]
- Ramos CG, Valdevenito R, Vergara I, et al. PCA3 sensitivity and specificity for prostate cancer detection in patients with abnormal PSA and/or suspicious digital rectal examination. First Latin American experience. Urol Oncol 2013;31:1522-6. [Crossref] [PubMed]
- Wei JT, Feng Z, Partin AW, et al. Can Urinary PCA3 Supplement PSA in the Early Detection of Prostate Cancer? J Clin Oncol 2014;32:4066-72. [Crossref] [PubMed]
- Salami SS, Schmidt F, Laxman B, et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol 2013;31:566-71. [Crossref] [PubMed]
- Leyten GHJM, Hessels D, Smit FP, et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin Cancer Res 2015;21:3061-70. [Crossref] [PubMed]
- Van Neste L, Hendriks RJ, Dijkstra S, et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur Urol 2016;70:740-8. [Crossref] [PubMed]
- Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 2009;100:1603-7. [Crossref] [PubMed]
- van der Vos KE, Balaj L, Skog J, et al. Brain Tumor Microvesicles: Insights into Intercellular Communication in the Nervous System. Cell Mol Neurobiol 2011;31:949-59. [Crossref] [PubMed]
- McKiernan J, Donovan MJ, O’Neill V, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol 2016;2:882-9. [Crossref] [PubMed]
- Stewart GD, Van Neste L, Delvenne P, et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: results of the MATLOC study. J Urol 2013;189:1110-6. [Crossref] [PubMed]
- Partin AW, Van Neste L, Klein EA, et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 2014;192:1081-7. [Crossref] [PubMed]
- Van Neste L, Partin AW, Stewart GD, Epstein JI, Harrison DJ, Van Criekinge W. Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. Prostate 2016;76:1078-87. [Crossref] [PubMed]
- Klein EA, Haddad Z, Yousefi K, et al. Decipher Genomic Classifier Measured on Prostate Biopsy Predicts Metastasis Risk. Urology 2016;90:148-52. [Crossref] [PubMed]
- Nguyen PL, Martin NE, Choeurng V, et al. Utilization of biopsy-based genomic classifier to predict distant metastasis after definitive radiation and short-course ADT for intermediate and high-risk prostate cancer. Prostate Cancer Prostatic Dis 2017;20:186-92. [Crossref] [PubMed]
- Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol 2014;66:550-60. [Crossref] [PubMed]
- Knezevic D, Goddard AD, Natraj N, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics 2013;14:690. [Crossref] [PubMed]
- Cullen J, Rosner IL, Brand TC, et al. A Biopsy-based 17-gene Genomic Prostate Score Predicts Recurrence After Radical Prostatectomy and Adverse Surgical Pathology in a Racially Diverse Population of Men with Clinically Low- and Intermediate-risk Prostate Cancer. Eur Urol 2015;68:123-31. [Crossref] [PubMed]
- Kornberg Z, Cowan JE, Masic S, et al. MP12-12 Genomic prostate score predicts progression in men on active surveillance. J Urol 2018;199. [Crossref]
- Shipitsin M, Small C, Giladi E, et al. Automated quantitative multiplex immunofluorescence in situ imaging identifies phospho-S6 and phospho-PRAS40 as predictive protein biomarkers for prostate cancer lethality. Proteome Sci 2014;12:40. [Crossref] [PubMed]
- Freedland SJ, Gerber L, Reid J, et al. Prognostic utility of cell cycle progression score in men with prostate cancer after primary external beam radiation therapy. Int J Radiat Oncol Biol Phys 2013;86:848-53. [Crossref] [PubMed]
- Blume-Jensen P, Berman DM, Rimm DL, et al. Development and clinical validation of an in situ biopsy-based multimarker assay for risk stratification in prostate cancer. Clin Cancer Res 2015;21:2591-600. [Crossref] [PubMed]
- Shipitsin M, Small C, Choudhury S, et al. Identification of proteomic biomarkers predicting prostate cancer aggressiveness and lethality despite biopsy-sampling error. Br J Cancer 2014;111:1201-12. [Crossref] [PubMed]
- Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 2013;8. [Crossref] [PubMed]
- Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol 2013;190:2047-53. [Crossref] [PubMed]
- Zhao SG, Chang S, Erho N, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol 2017;3:1663-72. [Crossref] [PubMed]
- Zhao SG, Chang SL, Spratt DE, et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: a matched, retrospective analysis. Lancet Oncol 2016;17:1612-20. [Crossref] [PubMed]
- Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245-55. [Crossref] [PubMed]
- Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol 2013;31:1428-34. [Crossref] [PubMed]
- Cuzick J, Berney DM, Fisher G, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer 2012;106:1095-9. [Crossref] [PubMed]
- Tosoian JJ, Chappidi MR, Bishoff JT, et al. Prognostic utility of biopsy-derived cell cycle progression score in patients with National Comprehensive Cancer Network low-risk prostate cancer undergoing radical prostatectomy: implications for treatment guidance. BJU Int 2017;120:808-14. [Crossref] [PubMed]
- Koch MO, Cho JS, Kaimakliotis HZ, et al. Use of the cell cycle progression (CCP) score for predicting systemic disease and response to radiation of biochemical recurrence. Cancer Biomark 2016;17:83-8. [Crossref] [PubMed]
- Cuzick J, Stone S, Fisher G, et al. Validation of an RNA cell cycle progression score for predicting death from prostate cancer in a conservatively managed needle biopsy cohort. Br J Cancer 2015;113:382-9. [Crossref] [PubMed]
- Chai H, Brown RE. Field effect in cancer-an update. Ann Clin Lab Sci 2009;39:331-7. [PubMed]
- Spratt DE, Yousefi K, Deheshi S, et al. Individual Patient-Level Meta-Analysis of the Performance of the Decipher Genomic Classifier in High-Risk Men After Prostatectomy to Predict Development of Metastatic Disease. J Clin Oncol 2017;35:1991-8. [Crossref] [PubMed]
- Spratt DE, Zhang J, Santiago-Jiménez M, et al. Development and Validation of a Novel Integrated Clinical-Genomic Risk Group Classification for Localized Prostate Cancer. J Clin Oncol 2018;36:581-90. [Crossref] [PubMed]
- Corcoran NM, Hovens CM, Hong MK, et al. Underestimation of Gleason score at prostate biopsy reflects sampling error in lower volume tumours. BJU Int 2012;109:660-4. [Crossref] [PubMed]
- Porten SP, Whitson JM, Cowan JE, et al. Changes in prostate cancer grade on serial biopsy in men undergoing active surveillance. J Clin Oncol 2011;29:2795-800. [Crossref] [PubMed]


