Eight tests for sperm DNA fragmentation and their roles in the clinic
In this issue, Agarwal and colleagues (1) have attempted the Herculean effort of sifting through hundreds of publications and several meta-analyses of those publications to answer a question that continues to plague clinicians in artificial reproduction technologies (ART) clinics. Simply stated, the question is “When is it appropriate to ask patients to spend more money to obtain a diagnosis of the status of DNA damage in the sperm as a tool for both the patient and clinician to decide on a course of treatment?”. They have also addressed a second, more difficult, question, although not as thoroughly because the data are just not there, which is, “If we do decide to assay sperm DNA damage, which test(s) would we use?” This problem continues to be debated in the literature with the general conclusion that there is a role for sperm DNA damage assessment but it is limited (2,3). Even though I am not a clinician, I have been privy to enough discussions on this issue with clinical colleagues to know that it is a troubling one. It takes time in an already crowded clinical day to explain to the patients what the test is and why they need it, and then why they should add this expense to their already high cost for treatment.
The issue for the clinician is making sense of the plethora of literature about how to apply the available tests for sperm DNA fragmentation (SDF) and which ones to use. The ART clinician may be excused for not having a good resource to which to refer when making this decision. The authors of this analysis entitled “Clinical utility of SDF testing: practice recommendations based on clinical scenarios” are all experienced clinical ART practitioners who have published extensively in this field. They have provided the ART clinician with an up-to-date summary of how eight different most commonly used SDF assays have been shown to be important for specific scenarios and have provided four recommendations for the application of SDF assays in the clinic. They also provide a level of confidence based on defined parameters of the evidence with A being the highest and C being the lowest. It is worth noting that none of the four recommendations received a Grade A level, and this reflects both the variety of the tests available, and the variation in study design of all the publications that are available for review.
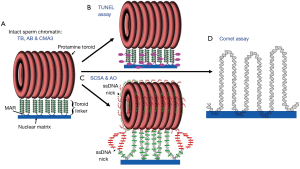
As a basic scientist interested in chromatin structure, it is difficult not to address the assays themselves and how they might measure different aspects of DNA packaging. We have proposed a model for mammalian sperm chromatin structure based on available evidence in the literature (4). Human sperm chromatin is divided into three major components: (I) 90% of the DNA is tightly compacted by protamines into toroids that each contain 20 to 50 kb of DNA (5), (II) about 10% of the DNA is packaged by histones (6) which we predict to be located at the sites that link the protamine toroids (7), and (III) nuclear matrix attachment sites (8,9). We have predicted that the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay primarily assays the protamine toroid linker DNA (Figure 1B). The sperm chromatin structure assay (SCSA) specifically removes histones, and probably some, but not all protamines, with acid extraction, thereby exposing histone bound DNA to denaturation required for the assay’s test of SDF. SCSA can assay the linker region and some damage in the protamine packaged toroids (Figure 1C) (10).This is probably due to the fact that the dye used to assay DNA damage in the SCSA test is much smaller than the enzyme used in the TUNEL assay, so it’s access to the DNA is not sterically hindered. The acridine orange (AO) test would have the same access to packaged DNA damage as the SCSA. The COMET assay is the only test that completely disrupts the both the histone and protamine packaging exposing naked DNA to the assay (Figure 1D) (11), however, this assay still depends on the attachment of the DNA to the nuclear matrix (12). CMA3 (13), TB or AB (14) staining do not appear to have a solution that removes histones, so these dyes must all assay the chromatin in its natural state. As such, one would expect the histone bound regions of the DNA to be more accessible to the dyes than the protamine bound regions. These are the regions of sperm chromatin that might also be expected to be more susceptible to DNA damaging events. Of the eight tests that Agarwal and colleagues (1) mentioned, the SCD is the only one for which we cannot guess the particular chromatin site that is affected because the extraction solution remains proprietary and undisclosed (15). However, given the halo appearance of the control, undamaged DNA, one might suggest that it is similar to the comet assay in exposing all the DNA to the test.
While it is clear that the eight different SDF assays that are discussed by Agarwal et al. (1) measure different aspects of sperm chromatin structure, much more work is need to clearly define exactly what parts of the chromatin structure each assay emphasizes. Further studies in this area will also advance our knowledge of normal and abnormal sperm chromatin structure and how specific sperm chromatin defects might influence reproductive outcomes.
Acknowledgements
Funding: The work was supported by NIH Grant No. GM103457 and HD060722.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Agarwal A, Roychoudhury S, Sharma R, et al. Diagnostic application of oxidation-reduction potential assay for measurement of oxidative stress: clinical utility in male factor infertility. Reprod Biomed Online 2017;34:48-57. [Crossref] [PubMed]
- Bach PV, Schlegel PN. Sperm DNA damage and its role in IVF and ICSI. Basic Clin Androl 2016;26:15. [Crossref] [PubMed]
- Bieniek JM, Lo KC. Recent advances in understanding & managing male infertility. F1000Res 2016;5:2756. [Crossref] [PubMed]
- Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod 2010;16:30-6. [Crossref] [PubMed]
- Hud NV, Downing KH. Cryoelectron microscopy of lambda phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc Natl Acad Sci U S A 2001;98:14925-30. [Crossref] [PubMed]
- Hammoud SS, Nix DA, Zhang H, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009;460:473-8. [PubMed]
- Sotolongo B, Lino E, Ward WS. Ability of hamster spermatozoa to digest their own DNA. Biol Reprod 2003;69:2029-35. [Crossref] [PubMed]
- Shaman JA, Yamauchi Y, Ward WS. The sperm nuclear matrix is required for paternal DNA replication. J Cell Biochem 2007;102:680-8. [Crossref] [PubMed]
- Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: a working model of potentiation. J Biol Chem 2004;279:51862-8. [Crossref] [PubMed]
- Gawecka JE, Boaz S, Kasperson K, et al. Luminal fluid of epididymis and vas deferens contributes to sperm chromatin fragmentation. Hum Reprod 2015;30:2725-36. [PubMed]
- Enciso M, Sarasa J, Agarwal A, et al. A two-tailed Comet assay for assessing DNA damage in spermatozoa. Reprod Biomed Online 2009;18:609-16. [Crossref] [PubMed]
- Ribas-Maynou J, Gawecka JE, Benet J, et al. Double-stranded DNA breaks hidden in the neutral Comet assay suggest a role of the sperm nuclear matrix in DNA integrity maintenance. Mol Hum Reprod 2014;20:330-40. [Crossref] [PubMed]
- Hosseinifar H, Yazdanikhah S, Modarresi T, et al. Correlation between sperm DNA fragmentation index and CMA3 positive spermatozoa in globozoospermic patients. Andrology 2015;3:526-31. [Crossref] [PubMed]
- Kim HS, Kang MJ, Kim SA, et al. The utility of sperm DNA damage assay using toluidine blue and aniline blue staining in routine semen analysis. Clin Exp Reprod Med 2013;40:23-8. [Crossref] [PubMed]
- García-Peiró A, Oliver-Bonet M, Navarro J, et al. Dynamics of sperm DNA fragmentation in patients carrying structurally rearranged chromosomes. Int J Androl 2011;34:e546-53. [Crossref] [PubMed]


