
ALDOB represents a potential prognostic biomarker for patients with clear cell renal cell carcinoma
Highlight box
Key findings
• ALDOB represents a potential prognostic biomarker for patients with clear cell renal cell carcinoma.
What is known and what is new?
• ALDOB might play controversial roles in multiple types of cancer, which could act as a cancer-promoting factor or a cancer-inhibiting factor depending on the subtype of the cancer.
• The expression level of ALDOB was significantly down-regulated in ccRCC compared to normal tissue, and the ALDOB expression level was noticeably correlated with T stage, M stage, and histologic grade of patients with ccRCC. Furthermore, ALODB was the independent predictor of OS, DSS, and PFS of ccRCC patients.
What is the implication, and what should change now?
• ALDOB might be a novel prognostic biomarker and therapeutic target for ccRCC patients. The nomograms with more prognostic predictive power were constructed to predict the OS, DSS, and PFS of ccRCC patients by integrating T stage, N stage, M stage, histologic grade, and the expression level of ALDOB of ccRCC patients. Based on these nomograms, it would be helpful to assess the clinical outcome of ccRCC more accurately and provide more personalized prognostic assessment strategy for ccRCC patients.
Introduction
As one of the common malignancies of the urinary system, the incidence of renal cell carcinoma (RCC) worldwide has shown a yearly increase in recent years (1-3). Among the three main types of RCC, clear cell renal cell carcinoma (ccRCC) is the main pathological type of RCC with a worse prognosis compared to papillary RCC and chromophobe RCC, accounting for 70% to 80% of all RCC (4,5). In the clinic, the majority of ccRCC are detected incidentally via noninvasive imaging investigation, and the classic triad of flank pain, visible haematuria, and palpable abdominal mass is rare, that generally correlates with aggressive disease (6). Currently, due to the lack of powerful diagnostic tools and prognostic evaluation systems other than imaging modalities, it is difficult for clinicians to early diagnose and precisely predict the prognosis of ccRCC. Therefore, the development of effective molecular biomarkers and therapeutic targets becomes essential to improve the prognosis of patients with ccRCC.
Aldolase B, encoded by the ALDOB gene, is mainly involved in the conversion of fructose 1-phosphate to dihydroxyacetone phosphate and glyceraldehyde (7). Increasing evidence confirms that ALDOB not only plays an important role in glycolysis and fructose metabolism, but is also aberrantly expressed in a variety of malignancies. And this abnormal expression pattern has been demonstrated to be highly associated with the clinicopathological features and prognosis of cancer patients (8-10). Tian et al. performed immunostaining to evaluate the expression level of the ALDOB protein in 172 patients with rectal adenocarcinomas treated with preoperative chemoradiotherapy followed by radical surgery (8). The results showed that overexpression of ALDOB was closely associated with clinicopathological factors of rectal adenocarcinoma patients, such as tumor advancement, lymphovascular invasion, and perineural invasion. And the survival analysis indicated that the high expression of ALDOB was significantly associated with worse disease-specific survival (DSS), which could be served as a prognostic biomarker for patients with rectal cancer (8). In addition, Li et al. used immunohistochemical staining to detect the protein expression level of ALDOB in 229 samples from patients with colorectal cancer who underwent primary tumor resection (9). The results showed that ALDOB overexpression was associated with poorer overall survival (OS) and disease-free survival (DFS) in patients with colorectal cancer. Further mechanistic study showed that ALDOB inactivation significantly inhibited the proliferation, migration, and invasive ability of colon cancer cells by participating in the regulation of the epithelial-mesenchymal transition (9). Contrarily, in a study of gastric cancer, ALDOB was found to be down-regulated in tumor tissues compared with adjacent nontumor tissues (10). And the expression of ALDOB was closely related to the tumor-invasion depth, lymph node metastasis, distant metastasis, and tumor stage. Combined with the results of survival analysis, they revealed that ALDOB was an independent prognostic factor for OS in gastric cancer patients, suggesting that ALDOB could act as a novel molecular marker for gastric cancer patients (10). Therefore, based on the previous studies, ALDOB might play controversial roles in different types of cancers, which could act as a cancer-promoting factor or a cancer-inhibiting factor depending on the subtype of the cancers.
Although extensive researches have been carried out on the role of ALDOB in other malignancies, few researches evaluated the expression level of and prognostic value of ALDOB in ccRCC patients. In addition, no single study has explored the role and value of ALDOB in ccRCC patients. Therefore, the main purpose of this study is to comprehensively explore the expression level, prognostic value, functional enrichment, immune infiltration, and N6-methyladenosine (m6A) modification of ALDOB in ccRCC patients by integrating multiple datasets. This study not only attempts to elucidate the role and value of ALDOB in ccRCC, but also targets to develop a new prognostic biomarker and therapeutic target for ccRCC patients. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-743/rc).
Methods
Download and processing of original expression profiles
The gene expression profiles of GSE53757 (11), GSE53000 (12), GSE36895 (13), GSE15641 (14), GSE66272 (15), GSE68417 (16), GSE40435 (17), GSE12606 (18), GSE46699 (19), and GSE11151 (20), were obtained from the Gene Expression Omnibus (GEO) database, totally including 439 ccRCC tissues and 337 normal kidney tissues (Table S1). The RNA sequencing (RNA-seq) data of 530 ccRCC tissues and 72 normal kidney tissues with corresponding clinicopathological features and survival data were downloaded and standardized from The Cancer Genome Atlas (TCGA) database (TCGA-KIRC dataset) (21). Moreover, the E-MTAB-1980 dataset, which contained 101 ccRCC patients with RNA-seq and clinical data, was downloaded from the ArrayExpress database (22). Therefore, a total of 1,070 ccRCC tissues and 409 normal tissues was enrolled in this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification and validation of differential expressed genes (DEGs)
Here, the DEGs between ccRCC and normal tissues were detected by the “limma” package in R software based on 439 ccRCC and 337 normal kidney tissues from ten GEO datasets. In this study, genes with P<0.05 and |log2(fold change)|>2 were considered as DEGs. The pivotal DEGs were acquired after the intersection of DEGs from ten GEO datasets by using the “UpSetR” package in R software. In addition, the expression levels of overlapping DEGs in 530 ccRCC and 72 normal kidney tissues from TCGA database were analyzed to further verify the results of the expression levels of pivotal DEGs obtained from the GEO datasets.
Prognostic value analysis of DEGs
The Kaplan-Meier survival curves were plotted, and the Log-Rank test was performed to assess the prognostic value of DEGs in ccRCC patients. And the primary outcomes of the survival analysis were OS, DSS, and progression-free survival (PFS). Here, hazard ratios (HRs) with 95% confidence intervals (CIs) were calculated to quantitatively estimate the prognostic value of DEGs. In addition, the univariate and multivariate Cox regression analyses were conducted to assess the influence on the survival of ccRCC patients within age, gender, T stage, N stage, M stage, AJCC stage, histologic grade, and DEGs expression level to further determine the independent predictors of OS, DSS, and PFS of ccRCC patients.
Expression analysis of ALDOB at mRNA and protein level
The mRNA expression level of ALDOB in ccRCC tissues and normal kidney tissues was analyzed and validated based on TCGA database as well as GEO database, respectively. Besides, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) (https://proteomics.cancer.gov/programs/cptac) database was employed to investigate the protein expression level of ALDOB in normal tissues and tumor tissues (23). Furthermore, the Human Protein Atlas (HPA) (http://www.proteinatlas.org/) database was utilized to visualize the immunohistochemistry images of ALDOB in ccRCC tissues and normal kidney tissues to verify the protein expression results of the CPTAC database (24). Finally, the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/) was used to predict the 3D protein structure of ALDOB (25).
Clinicopathological analysis of ALDOB
Based on the expression level of ALDOB and the clinicopathological data of ccRCC patients from TCGA database, the relationship between the mRNA expression level of ALDOB and T-stage, N-stage, M-stage, and pathological grade of ccRCC patients were determined. Furthermore, the logistic regression analyses were also used to validate the association of ALDOB and the clinicopathological characteristics of ccRCC patients from TCGA-KIRC dataset.
Construction and validation of nomograms
Based on the results of the Cox logistic regression analysis, the prognostic predictors, including T stage, N stage, M stage, AJCC stage, pathological grade, and the expression level of ALDOB in ccRCC patients, were employed to construct the nomograms of 1-year, 3-year, and 5-year OS, DSS, and PFS of ccRCC patients from TCGA-KIRC dataset. Besides, the concordance index (C-index) was calculated by 1,000 bootstrap resamples, and the calibration curves were utilized to evaluate the predictive accuracy and reliability of the nomograms.
Protein-protein interaction network and functional enrichment analysis
In this work, a protein-protein interaction (PPI) network of ALDOB was built to predict the functional partners of ALDOB based on the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (26). Additionally, the Gene Ontology (GO) analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of ALDOB and its correlated genes were performed to reveal the potential functions and possible signaling pathways of ALDOB in ccRCC.
Immune infiltration analysis
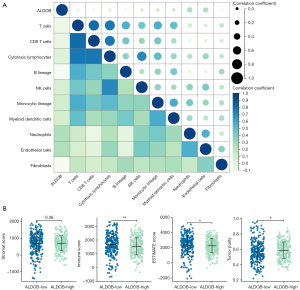
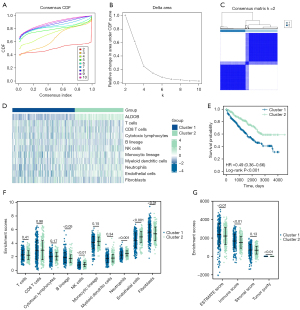
Based on the “MCP-Counter” algorithm, this study analyzed the infiltration abundance of eight types of immune cells (T cells, CD8 T cells, cytotoxic lymphocytes, B lineage cells, natural killer cells, monocyte lineage, myeloid dendritic cells, and neutrophils) and two types of stromal cells (endothelial cells and fibroblasts) in ccRCC patients from the TCGA-KIRC dataset to explore the correlation between the expression level of ALDOB and the abundance of immune cells and stromal cells in the tumor microenvironment (TME) (27). Moreover, the “Estimate” package in R software was employed to calculate the Immune Score, Stromal Score, ESTIMATE Score, and Tumor Purity that respectively represented the immune component, stromal component, and the tumor purity for each ccRCC sample (28). Then, in order to determine whether ALDOB and ten types of immune cells and stromal cells could distinguish ccRCC patients, the “ConsensusClusterPlus” package in R software was used to perform consensus clustering analysis for subtyping the ccRCC patients from the TCGA-KIRC dataset. And the consensus matrix k value denoted the number of clusters. At the same time, the Kaplan-Meier survival curves and the Log-Rank test were also performed to compare the prognosis between the two clusters. Finally, the “MCP-Counter” and “Estimate” algorithm were also used to evaluate the relative abundance of immune cells and TME within two different clusters.
m6A methylation analysis of ALDOB
Initially, the correlation between ALDOB and a total of 23 types of m6A regulators, including eight writers (METTL3, METTL14, RBM15B, RBM15, VIRMA, WTAP, CBLL1, and ZC3H13), two erasers (FTO and ALKBH5), and 13 readers (YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, HNRNPC, HNRNPA2B1, IGF2BP1, IGF2BP2, IGF2BP3, LRPPRC, FMR1, and ELAVL1) was examined to explore the potential mechanism for the abnormal expression pattern of ALDOB in ccRCC. Then, the least absolute shrinkage and selection operator (LASSO) regression was conducted for selecting and identifying a panel of genes within ALDOB and 23 m6A modulators via “glmnet and survival” package in R software based on TCGA-KIRC dataset. Next, the multivariate Cox regression analysis was applied to establish a prognostic signature and calculate Risk Scores for each ccRCC patients in TCGA-KIRC dataset. Afterward, 530 ccRCC patients from TCGA-KIRC dataset were classified into low-risk group and high-risk group according to the optimal cut-off value of the risk scores. Subsequently, the Kaplan-Meier survival curve was performed with the Log-Rank test to compare the prognosis between the two risk groups. Lastly, the time-dependent receiver operating characteristic (ROC) curve and the area under curve (AUC) values were used to analyze the sensitivity and specificity in evaluating the accuracy of the model that this study constructed.
Statistical analysis
The Mann-Whitney U test and Student’s t-test was used to compare the differences between two groups. The Kruskal-Wallis H test and Dunn’s test were performed to evaluate the relationships between clinicopathological features and the expression level of ALDOB. The Kaplan-Meier curves were depicted to assess the prognostic value of genes in ccRCC patients and the Log-Rank test was performed to assess the survival differences between two groups. The univariate and multivariate Cox regression analysis were used to identify the independent prognostic predictors. The Pearson correlation test was conducted for the correlation analysis. In this study, the statistical analysis was performed using R software (version 4.2.0). And the figures were drawn with R software (version 4.2.0) and the corresponding online databases. Statistical significance was set at the P value <0.05 for all the analyses. The flow diagram of this study is presented in Figure 1.
Results
Identification and validation of DEGs
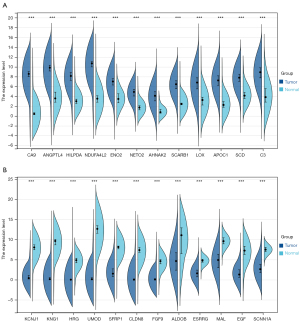
According to the set criteria of DEGs, this study analyzed the DEGs and depicted the volcano plots in each GEO dataset (Figure S1). As a result, a total of 24 DEGs was screened and determined after taking the intersection of DEGs of ten GEO datasets (Figure S2). Among them, 12 DEGs were up-regulated in ccRCC tissues, including CA9, ANGPTL4, HILPDA, NDUFA4L2, ENO2, NETO2, AHNAK2, SCARB1, LOX, APOC1, SCD, and C3. And 12 DEGs were expressed at significantly lower levels in ccRCC tissues, including KCNJ1, KNG1, HRG, UMOD, SFRP1, CLDN8, FGF9, ALDOB, ESRRG, MAL, EGF, and SCNN1A (Figure S2). In addition, this study verified the expression levels of 24 DEGs in 530 ccRCC tissues and 72 normal kidney tissues from the TCGA-KIRC dataset. As expected, the expression levels of 24 DEGs were same as those in ten GEO datasets (Figure 2).
Prognostic value of DEGs in ccRCC patients
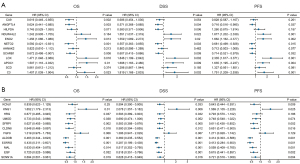
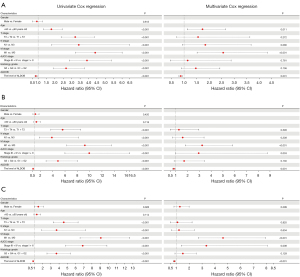
In order to explore the prognostic value of DEGs in ccRCC patients, the univariate Cox regression analysis was first employed to perform the survival analysis based on the clinical data from TCGA-KIRC dataset. The results showed that among the 12 up-regulated genes, the expression levels of ANGPTL4, ENO2, NETO2, SCARB1, LOX, APOC1, and C3 were associated with OS, DSS, and PFS of ccRCC patients (Figure 3A). The ccRCC patients with high expression level of ANGPTL4, NETO2, and SCARB1 experienced a better prognosis, and ccRCC patients with high expression of ENO2, LOX, APOC1, and C3 genes had a significantly shorter survival time (Figure 3A). Moreover, among the 12 down-regulated genes, the expression levels of KNG1, SFRP1, CLDN8, ALDOB, ESRRG, MAL, EGF, and SCNN1A were correlated with OS, DSS, and PFS of ccRCC patients (Figure 3B). And the prognosis of ccRCC patients with high expression levels of SFRP1, CLDN8, ALDOB, ESRRG, MAL, EGF, and SCNN1A were better (Figure 3B). Subsequently, the multivariate Cox regression analyses were conducted to further identify the independent predictors of OS, DSS, and PFS in ccRCC patients. The forest plots showed that, only the expression level of ALDOB was closely related to OS (HR =0.866, 95% CI: 0.794–0.945, P=0.001), DSS (HR =0.830, 95% CI: 0.741–0.931, P=0.001), and PFS (HR =0.838, 95% CI: 0.764–0.919, P<0.001) in ccRCC patients (Figure 4). And the ccRCC patients with higher expression level of ALDOB experienced a better prognosis, while those with lower expression level of ALDOB had a significantly worse prognosis. Furthermore, the prognostic value of ALDOB in ccRCC patients was also validated by multivariate Cox regression analysis in an independent cohort (E-MTAB-1980 dataset) (Figure S3). Therefore, ALDOB was identified as an independent factor for prognosis of ccRCC patients (Figure 4).
The expression level of ALDOB in ccRCC patients
In this analysis, the expression level of the ALDOB mRNA was investigated based on the ten GEO datasets. The results suggested that the mRNA expression level of ALDOB was significantly down-regulated in ccRCC tissues compared to that in the normal tissues (Figure 5A). Furthermore, the analysis of the mRNA expression level of ALDOB from TCGA-KIRC dataset indicated the similar expression patterns in the ccRCC and normal samples (Figure 5A), further validating the findings of the analysis performed using the GEO datasets. In addition to analyzing the expression level of ALDOB at mRNA level, the expression of ALDOB at the protein level was also analyzed by using the CPTAC database and the HPA database. The results show that the protein expression level of ALDOB in the 110 ccRCC samples was significantly lower than that in the 84 normal tissues from the CPTAC database (Figure 5B). Moreover, a lower protein expression level of ALDOB protein was also found in the ccRCC tissues than that in the normal tissues, as visualized by the representative immunohistochemical images from the HPA database (Figure 5C). The 3D structure of ALDOB predicted by the AlphaFold database is shown in Figure 5D. In summary, these results indicated that ALDOB is down-regulated in ccRCC at both transcriptional and translational levels.
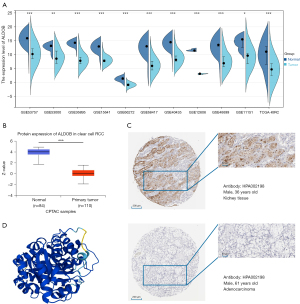
Relationship between the expression level of ALDOB and the clinicopathological features of ccRCC patients
To determine the relationship between the expression of ALDOB and the clinicopathological features of ccRCC patients, the characteristics of the ccRCC patients from the TCGA-KIRC dataset were further analyzed. The results from the logistic regression analyses showed that the expression level of ALDOB was also closely related to the T stage, AJCC stage, and histologic grade of ccRCC patients (Table S2). Furthermore, the results showed a decreasing trend of ALDOB expression with an upgrade of T stage, M stage, AJCC stage, and histologic grade of ccRCC patients (Figure 6). And the ccRCC patients with T1 stage, metastasis, AJCC stage 1, and histologic grade 1 were found to have the higher expression level of ALDOB (Figures 6B-6E). To summarize, these results showed that ALDOB might be associated with tumor development and progression in ccRCC patients.
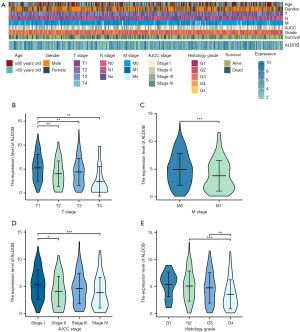
Construction and validation of nomograms
Based on the results of Cox logistic regression analyses, the prognostic predictors, including T stage, N stage, M stage, AJCC stage, histologic grade, and ALDOB, were used to establish the nomograms of 1-year, 3-year, and 5-year OS, DSS, and PFS of ccRCC patients from TCGA-KIRC dataset. These nomograms were used to provide clinically quantitative tools to predict the prognosis of the patients with ccRCC (Figure 7A-7C). The C-indices of the predicted OS, DSS, and PFS were 0.764 (0.737–0.790), 0.860 (0.839–0.881), and 0.823 (0.801–0.846), respectively, suggesting the excellent prognostic ability for the ALDOB-based nomograms. Moreover, the calibration plots showed good consistency between the predicted survival and observation survival of OS, DSS, and PFS (Figures 7D-7F), demonstrating that these nomograms performed well and were ideal models for predicting the prognosis for ccRCC patients. Furthermore, the nomogram and calibration plot of 1-year, 3-year, and 5-year OS of ccRCC patients were also established in the external cohort (E-MTAB-1980 dataset), which validated the good prognostic ability for the ALDOB-based model (Figure S3). Overall, ALDOB might be a novel prognostic biomarker for predicting the prognosis of ccRCC patients.
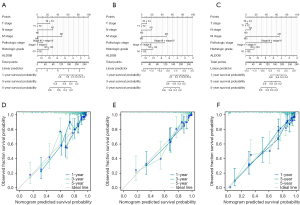
Functional enrichment analysis and protein-protein interaction network
Firstly, the correlation analysis was performed to explore the potential related genes that might interact with ALDOB. The correlation circle plot and heatmap showed that the top ten positively correlated genes that interacted with ALDOB were G6PC, CYP4A11, FUT6, PKLR, SLC22A6, AGMAT, CYP4A22, SLC22A7, AKR7A3, and SLC22A12 (Figure 8A,8B). Then, the GO and KEGG analysis pathway analysis of ALDOB and its correlated genes was carried out to reveal the potential functions of ALDOB in ccRCC. The results showed that in ccRCC, ALDOB was mainly involved and participated in the small molecule catabolic process (Biological Process), apical part of cell (Cellular Component), organic anion transmembrane transporter activity (Molecular Function), and PPAR signaling pathway (KEGG Pathway) (Figure 8C). Finally, by the order of the predicted scores, a PPI network of ALDOB was built based on the STRING database. And the result showed that the predicted interactive partners of ALDOB were TPI1, FBP1, PFKL, FBP2, and PFKP (Figure 8D).
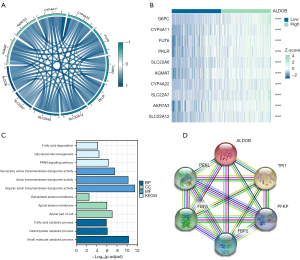
Immune infiltration analysis of ALDOB
Based on the “MCP-Counter” algorithm, this study firstly analyzed the infiltration abundance of eight types of immune cells and two types of stromal cells in ccRCC patients from TCGA-KIRC dataset. We observed that the expression level of ALDOB was correlated with the abundance of most of immune cells and stromal cells (Figure 9A). We next calculated the immune score, stromal score, estimate score, and tumor purity of ccRCC patients. The results showed that the immune score and the estimate score were higher in the ALDOB-low cohort than in the ALDOB-high cohort (Figure 9B). However, the tumor purity showed the opposite difference (Figure 9B). Therefore, the above results showed the significant correlation between ALDOB and the immune infiltration in ccRCC. In order to determine whether ALDOB and ten types of immune cells and stromal cells could distinguish ccRCC patients, we next performed consensus clustering analysis. The results showed that ALDOB and immune cells could classify the ccRCC patients from TCGA-KIRC dataset into two distinct clusters (Figure 10A-10C). And the heatmap demonstrated the specific expression level of ALDOB and the infiltration abundance of immune cells and stromal cells among two different clusters (Figure 10D). We also observed that the ccRCC patients in the cluster 1 were more prone to experience a shorter OS than the patients in the cluster 2 (HR =0.490, 95% CI: 0.360–0.660, P<0.001) (Figure 10E). Moreover, the B lineage, fibroblasts, estimate scores, and immune scores were elevated in the cluster 1, whereas the ccRCC patients in the cluster 2 had significantly higher levels of the natural killer (NK) cells, neutrophils, endothelial cells, and tumor purity scores (Figure 10F,10G). Taken together, these results confirmed that ALDOB strongly affects the immune infiltration in ccRCC.
m6A methylation analysis of ALDOB
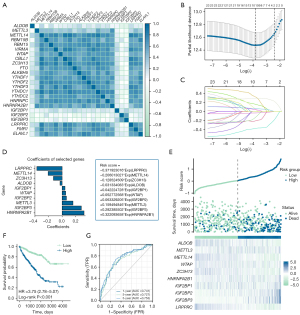
The correlation between the expression levels of ALDOB and 23 m6A regulators was first investigated. The correlation heatmap showed that the expression level of ALDOB was significantly correlated with the expression levels of METTL14 (r=0.25, P<0.001), ALKBH5 (r=0.24, P<0.001), YTHDC1 (r=0.22, P<0.001), RBM15B (r=0.21, P<0.001), LRPPRC (r=0.20, P<0.001), IGF2BP2 (r=−0.31, P<0.001), and IGF2BP3 (r=−0.29, P<0.001) (Figure 11A). Then, the LASSO regression analysis with 10-fold cross-validation was carried out to identify the prognostic factors that were significantly correlated with the prognosis of ccRCC patients based on ALDOB and 23 m6A regulators (Figure 11B,11C). As a result, a total of ten genes, including HNRNPA2B1, IGF2BP3, METTL3, IGF2BP2, WTAP, IGF2BP1, ALDOB, ZC3H13, METTL14, and LRPPRC, were identified and enrolled through the LASSO regression analysis. Therefore, this study established the prognostic model and calculated the risk scores for each ccRCC patient based on these ten genes (Figure 11D). The risk factor diagram showed the difference in the distribution of the expression level of ten genes in the high-risk cohorts and low-risk cohorts (Figure 11E). Subsequently, the results of the survival analysis indicated that, compared to the ccRCC patients in high-risk groups, the ccRCC patients in low-risk groups experienced a better OS (HR =3.750, 95% CI: 2.780–5.070, P<0.001) (Figure 11F). Finally, the AUC value calculated based on the time-dependent ROC curve was high, indicating the prognostic accuracy of the model that this study constructed (Figure 11G).
Discussion
For localised ccRCC patients, surgery is still the only curative treatment (4). However, due to 20–30% of ccRCC patients experience tumor recurrence and metastasis after surgery, there is an urgent need to diagnose, monitor, and manage this cancer (29,30). Currently, apart from imaging investigations, the reliable and effective molecular biomarkers for ccRCC that suitable for clinical application have not yet been identified, and the effective risk stratification and screening systems have not been established. The lack of early diagnostic tools has added much pressure and difficulties to the early detection and early treatment of ccRCC. Consequently, development of effective molecular biomarkers is essential to improve the clinical management and reduce the mortality of ccRCC patients.
Initially, this study was devoted to screen and identify the DEGs in ccRCC. Through the intersection of DEGs from ten GEO datasets, a total of 24 DEGs was determined, including twelve up-regulated genes and twelve down-regulated genes. Besides, the expression levels of the 24 DEGs in ccRCC tissues and normal kidney tissues were verified using the TCGA-KIRC datasets. Subsequently, the results of survival analysis showed that the expression levels of 15 genes were markedly correlated with OS, DSS, and PFS of ccRCC patients. On the basis of univariate Cox regression analyses, the results of multivariate Cox regression analysis showed that, among the 24 DEGs, only the expression level of ALDOB was an independent predictor of OS, DSS, and PFS of ccRCC patients. Therefore, ALDOB was identified as a pivotal gene in ccRCC.
Metabolic patterns of cellular processes have been reported to be associated with a variety of tumorigenesis (31). Recent developments in the field of cancers have led to an interest in the changes in the metabolic pathways that control tumor energy and biosynthesis, known as metabolic reprogramming, which was considered to be a core hallmark of cancer (32,33). Moreover, the alterations of the known renal cell carcinoma genes, such as VHL, is closely related to the reprogramming of metabolic pathways (34). A number of researchers have reported that the development of ccRCC is usually accompanied by the metabolic reprogramming of glycolysis, fatty acid metabolism, and the tricarboxylic acid cycle (35-37). Therefore, glycolysis plays a vital role in the metabolic network. The progress of glycolysis usually consists of ten main steps, involving the participation of multiple enzymes (38). The members of the aldolase family are the fourth enzyme in the progress of glycolysis, that consists of ALDOB, ALDOB, and ALDOC, encoding by three different genes (7). Aldolase B, also known as fructose bisphosphate aldolase B, is encoded by the ALDOB gene and is mainly expressed in the liver and kidney tissues (7). In vivo, aldolase B catalyzes the conversion of fructose 1-phosphate to dihydroxyacetone phosphate and glyceraldehyde (39). It has been demonstrated that ALDOB not only plays an important role in glucose and fructose metabolism, but is also differentially expressed in a variety of malignancies, such as rectal adenocarcinoma, colorectal cancer, and gastric cancer (8-10). However, few studies have been able to draw on any systematic research into the role of ALDOB in ccRCC patients. Therefore, the aim of this study was to clarify the expression level, prognostic value, functional enrichment, immune infiltration, and m6A modification of ALDOB in ccRCC patients. In this study, we first observed that the mRNA and protein expression level of ALDOB was down-regulated in ccRCC patients. Furthermore, the results showed a decreasing trend of ALDOB expression with an upgrade of T stage, M stage, AJCC stage, and histologic grade of ccRCC patients, suggesting that ALDOB was strongly correlated with the clinicopathological factors of ccRCC patients and might strongly influence tumor development and progression in ccRCC.
With continuous researches, several studies evaluated the prognostic value of ALDOB in a variety of malignancies and delved into the molecular mechanisms of ALDOB in the development of cancers. In a prostate cancer study, Xia et al. showed that the ALDOB, as an independent risk factor for metastasis-free survival of prostate cancer patients, was significantly associated with the pathological grade and tumor stage of patients with prostate cancer (40). And the prostate cancer patients in high-risk group exhibited therapeutic resistance and immunosuppression. Further experiments in vitro showed that the expression of ALDOB increased significantly under the hypoxic conditions and promoted the proliferation and invasion ability of prostate cancer cells (40). Several studies have confirmed that ALDOB could also regulate the progression of malignancies by participating in various important signaling pathways, including PI3K/AKT/mTOR signaling pathway, GSK-3β signaling pathway, and Wnt signaling pathway (41-43). In addition, it was shown that the expression level of ALDOB was up-regulated in the liver metastasis tumors of colon cancer, providing sufficient energy supply to tumor cells by enhancing fructose metabolism (44). And the loss of ALDOB decreased the proliferative rates of tumor cells in the liver. Interestingly, this study also showed that reducing dietary fructose significantly reduces liver metastatic growth (44). Also, a research by Li et al. pointed out that ALDOB could suppress hepatocellular carcinogenesis by directly binding and inhibiting the activity of glucose-6-phosphate dehydrogenase, the rate-limiting enzyme in the pentose phosphate pathway, revealing a new mode of metabolic reprogramming in hepatocellular carcinogenesis due to the loss of ALDOB (45). Based on the above research background, this study further discussed the prognostic value of ALDOB in ccRCC patients. The results of multivariate Cox regression analysis showed that among the 24 DEGs, only ALDOB was an independent prognostic factor for OS, DSS, and PFS in ccRCC patients, and the ccRCC patients with higher expression level of ALDOB experienced a better prognosis, partly being attributed to the fact that the ccRCC patients with higher expression level of ALDOB had lower T stage, N stage, M stage, and histologic grade. Previously, some scholars also explored the mechanism of ALDOB in ccRCC. Wang et al. found that the accumulation of fructose 1, 6-bisphosphate in cancer cells by the down-regulation of ALDOB maintained the redox homeostasis and promoted ccRCC growth (46). What’s more, in a genetic assessment on frozen tissue samples of metastatic ccRCC, Nouhaud et al. used quantitative multiplex polymerase chain reaction of short fluorescent fragment method to detect the gene copy number variations on the 14 genes including ALDOB (47). The results showed that the most frequent losses were located on ALDOB, and the loss of ALDOB was associated with higher histologic grade and worse prognosis of metastatic ccRCC patients (47). In summary, ALDOB could play an important role in the development of ccRCC and influence the prognosis of ccRCC patients through multiple pathways and mechanisms.
Presently, the prognosis of ccRCC patients treated with radical nephrectomy or partial nephrectomy always depends on the anatomical, histological, clinical, and molecular factors (4). Among them, anatomical factors are reflected in the TNM classification, providing the most reliable information. And histological factors include RCC subtype, tumor grade, sarcomatoid features, vascular invasion, tumor necrosis, invasion of the collecting system, and perirenal fat. For the patients with localized and locally advanced ccRCC, the commonly used prognostic evaluation systems are the University of California, Los Angeles, Integrated Staging System (UISS) and Stage, Size, Grade, and Necrosis (SSIGN) scores (48,49), while for advanced or metastatic ccRCC, the prognostic risk models are the International Metastatic RCC Database Consortium (IMDC) and the Memorial Sloan Kettering Cancer Center (MSKCC) scoring systems (50). The results of this study showed that ALDOB was an independent prognostic factor in the prognosis of ccRCC patients. Therefore, in this study, the nomograms with more prognostic predictive power were developed to predict the OS, DSS, and PFS of ccRCC patients by integrating T stage, N stage, M stage, histologic grade, and the expression level of ALDOB of ccRCC patients. Based on these nomograms, it would be helpful to more accurately assess the clinical outcome of ccRCC and provide more personalized prognostic assessment strategy for ccRCC patients.
Subsequently, to determine the potential functions and interactive partners of ALDOB in ccRCC, this study first analyzed the potential related genes that might interact with ALDOB in ccRCC patients. Next, the GO and KEGG analysis of ALDOB and its related genes were performed. The results showed that ALDOB and its related genes are mainly involved in the metabolism and metabolic pathways of multiple substances, including small molecule catabolic process, glycolysis, gluconeogenesis, and fatty acid degradation. Therefore, this study confirmed that the aberrant expression of ALDOB is potentially involved in the metabolic reprogramming in ccRCC patients and is closely associated with reprogramming of diverse metabolic pathways and metabolic networks in ccRCC. Besides, the results of PPI network analysis suggested that the predicted functional partners of ALDOB were TPI1, FBP1, PFKL, FBP2, and PFKP. TPI1 gene codes triosephosphate isomerase 1, that catalyzes the interconversion between dihydroxyacetone phosphate and D-glyceraldehyde-3-phosphate in glycolysis and gluconeogenesis (51). FBP1 gene codes fructose-bisphosphatase 1, that catalyzes the hydrolysis of fructose 1,6-bisphosphate to fructose 6-phosphate in the presence of divalent cations, acting as a rate-limiting enzyme in gluconeogenesis (52). PFKL gene codes phosphofructokinase, that catalyzes the phosphorylation of D-fructose 6-phosphate to fructose 1,6-bisphosphate by ATP, which is the first committing step of glycolysis (53). FBP2 gene codes fructose-bisphosphatase 2, that catalyzes the hydrolysis of fructose 1,6-bisphosphate to fructose 6-phosphate in the presence of divalent cations, and probably participates in glycogen synthesis from carbohydrate precursors, such as lactate (54). PFKP gene codes phosphofructokinase, that catalyzes the phosphorylation of D-fructose 6-phosphate to fructose 1,6-bisphosphate by ATP, which is the first committing step of glycolysis (55). Therefore, ALDOB and its functional partners are mainly involved in a variety of pathways such as fructose metabolism and glycolysis. Previous studies have revealed that the metabolic reprogramming in ccRCC is mainly focused on the glucose metabolism, tricarboxylic acid cycle, and fatty acid oxidation and synthesis (35-37). The findings of this study implied that the disorders of fructose metabolism might also exist in ccRCC and ALDOB-mediated fructose metabolism might drive metabolic reprogramming in ccRCC, which provides a potential direction for further research.
The TME refers to the cellular environment in which the tumor exists (56). Numerous studies have confirmed that TME plays an important role in tumor development, adaptation, and pathogenesis (57-59). Therefore, understanding how to regulate the balance of TME within and between different tumor-supporting and tumor-suppressing infiltrating immune cells will hopefully lead to potentially more effective therapy for patients with cancers. In addition to tumor cells, the TME also includes immune cells, stromal cells, blood vessels, extracellular matrix, and other signaling molecules (60). Accordingly, in this study, we elucidated the infiltration abundance of immune cells and stromal cells in ccRCC patients and determined the correlation between ALDOB and these cells. The result indicated that the expression level of ALDOB was correlated with the abundance of most immune cells and stromal cells. In terms of immune scores, we observed ccRCC patients in the ALDOB-low expression group had elevated immune scores than those in ALDOB-high expression group. These results implicated that ALDOB might regulate the infiltrating abundance of immune cells and stromal cells in TME, thus exerting a potential influence on the prognosis of ccRCC patients. To confirm this hypothesis, the consensus clustering analysis was further performed based on the expression level of ALDOB and the abundance of immune cells and stromal cells to subtype the ccRCC patients. The results indicated that the ccRCC patients in the cluster 1 had the higher abundance of fibroblasts. In the physiological conditions, the primary function of fibroblasts is to support tissue integrity and participate in the wound healing (57). However, in the cancerous conditions, dysregulated cancer associated fibroblasts (CAFs) could promote the inflammatory response by producing cytokines and chemokines to recruit immune cells (57). On the other hand, CAFs could inhibit the antitumor activity of antigen-presenting cells by down-regulating the co-stimulatory molecules in dendritic cells (DCs) and stimulating the differentiation of immature DCs to regulatory DCs with immunosuppressive activity (57). Thus, CAFs could directly or indirectly contribute to the tumor development by driving immune suppression. Therefore, the ccRCC patients in cluster 1 were more prone to poor prognosis, partly attributed to the higher abundance of fibroblasts. In contrast, we observed a higher abundance of NK cells in the cluster 2 patients. As a central population of innate lymphoid cells, NK cells play a crucial role in triggering host immune responses against the tumor growth, and the immunosurveillance of solid tumors (61). For many patients with solid tumors, the high level of tumor infiltrating NK cells in TME predicts a good prognosis (61). Similarly, the findings of this study revealed that the ccRCC patients in the cluster 2 experienced a better prognosis, possibly due to the higher abundance of NK cells in these patients. Taken together, this study suggested that ALDOB might involve in regulating the infiltration abundance of immune cells and stromal cells to change the immune components of the TME, which in turn affects the prognosis of ccRCC patients.
m6A modification is the most prevalent, abundant, and conserved post-transcriptional modification to control the gene expression in the eukaryotic cells (62). As one of the most common reversibly epigenetic modification, m6A modification is widespread on mRNA and noncoding RNAs, that having important effects on the stability, localization, translation, splicing, and transport of RNAs (63). Currently, numerous studies have shown that the abnormalities of m6A modification are found in the various malignancies, and are strongly associated with the progression, metastasis, prognosis, and drug resistance of patients with tumors (63-65). Generally, there are three types of proteins that modulate m6A modifications, namely writers, erasers, and readers. In some types of cancers, these three types of m6A regulatory proteins are commonly dysregulated and could involve in different signaling pathways to influence the progression and prognosis of cancer patients (65). Therefore, in this study, the correlation between ALDOB and 23 m6A regulators was first investigated. The results showed that ALDOB was closely associated with multiple m6A regulators. Then, a risk score formula based on the ALDOB and nine m6A regulators was constructed after the LASSO regression analysis. The results showed that the models we constructed could effectively predict the prognosis of ccRCC patients, and ccRCC patients in the high-risk group were more prone to poorer OS than those with lower risk scores. In summary, this study revealed the potential correlation between ALDOB and m6A modifications, and highlighted the importance of the prognostic impact of ALDOB and m6A modulators on ccRCC patients.
Although this study aimed to undertake a comprehensive analysis of the expression level, prognostic value, functional enrichment, immune infiltration, and m6A modification of ALDOB in ccRCC, it still inevitably comes with limitations. First, this study included as many datasets as possible to fully demonstrate the expression pattern of ALDOB. However, further studies based on the prospective clinical studies and vitro cellular experiments are needed to validate the expression level and explore the specific mechanism of ALDOB in ccRCC. Second, this study investigated the potential correlation between ALDOB and TME and m6A modification. However, these hypotheses are required to be confirmed by the basic experiments to clarify the possible regulatory mechanisms. Finally, despite the fact that this study included samples from 1,479 tissues from ccRCC patients, the conclusions of this study were based on bioinformatic analysis and retrospective publicly available databases, making the conclusions less convincing. Therefore, more genomic and metabolomics studies are needed to further confirm the role of ALDOB in fructose metabolism and metabolic reprogramming in ccRCC. It would be the goal and direction of our future research.
Conclusions
Taken together, the findings of this study showed that the down-regulated ALDOB in ccRCC patients was closely associated with the clinicopathological features, poor prognosis, and TME in patients with ccRCC. Therefore, ALDOB might be a novel prognostic biomarker and therapeutic target for ccRCC patients. This study not only revealed that ALDOB-mediated fructose metabolism might drive metabolic reprogramming in ccRCC, but also suggested a new perspective for the precision medicine in ccRCC patients.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant No. 81872078), the Natural Science Foundation of Tianjin (grant No. 21JCYBJC0143), the Central Government Guides the Construction of Scientific and Technological Innovation Bases for Local Scientific and Technological Development funds (grant No. 22ZYJDSY00010), the Beijing Bethune Charitable Foundation (Grant No. mnz1202029), and the Scientific Research Funding Project of Returned Overseas Scholars of Shanxi Province (grant No. 2021-160).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-743/rc
Peer Review File: Available at https://tau.amegroups.com/article/view/10.21037/tau-22-743/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-743/coif). YN serves as an unpaid editorial board member of Translational Andrology and Urology. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Bukavina L, Bensalah K, Bray F, et al. Epidemiology of Renal Cell Carcinoma: 2022 Update. Eur Urol 2022;82:529-42. [Crossref] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer 2018;103:356-87. [Crossref] [PubMed]
- Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2022 Update. Eur Urol 2022;82:399-410. [Crossref] [PubMed]
- Moch H, Amin MB, Berney DM, et al. The 2022 World Health Organization Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur Urol 2022;82:458-68. [Crossref] [PubMed]
- Campbell SC, Clark PE, Chang SS, et al. Renal Mass and Localized Renal Cancer: Evaluation, Management, and Follow-Up: AUA Guideline: Part I. J Urol 2021;206:199-208. [Crossref] [PubMed]
- Chang YC, Yang YC, Tien CP, et al. Roles of Aldolase Family Genes in Human Cancers and Diseases. Trends Endocrinol Metab 2018;29:549-59. [Crossref] [PubMed]
- Tian YF, Hsieh PL, Lin CY, et al. High Expression of Aldolase B Confers a Poor Prognosis for Rectal Cancer Patients Receiving Neoadjuvant Chemoradiotherapy. J Cancer 2017;8:1197-204. [Crossref] [PubMed]
- Li Q, Li Y, Xu J, et al. Aldolase B Overexpression is Associated with Poor Prognosis and Promotes Tumor Progression by Epithelial-Mesenchymal Transition in Colorectal Adenocarcinoma. Cell Physiol Biochem 2017;42:397-406. [Crossref] [PubMed]
- He J, Jin Y, Chen Y, et al. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther 2016;9:6099-109. [Crossref] [PubMed]
- von Roemeling CA, Radisky DC, Marlow LA, et al. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res 2014;74:4796-810. [Crossref] [PubMed]
- Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225-33. [Crossref] [PubMed]
- Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 2012;44:751-9. [Crossref] [PubMed]
- Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res 2005;11:5730-9. [Crossref] [PubMed]
- Wotschofsky Z, Gummlich L, Liep J, et al. Integrated microRNA and mRNA Signature Associated with the Transition from the Locally Confined to the Metastasized Clear Cell Renal Cell Carcinoma Exemplified by miR-146-5p. PLoS One 2016;11:e0148746. [Crossref] [PubMed]
- Thibodeau BJ, Fulton M, Fortier LE, et al. Characterization of clear cell renal cell carcinoma by gene expression profiling. Urol Oncol 2016;34:168.e1-9. [Crossref] [PubMed]
- Wozniak MB, Le Calvez-Kelm F, Abedi-Ardekani B, et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS One 2013;8:e57886. [Crossref] [PubMed]
- Stickel JS, Weinzierl AO, Hillen N, et al. HLA ligand profiles of primary renal cell carcinoma maintained in metastases. Cancer Immunol Immunother 2009;58:1407-17. [Crossref] [PubMed]
- Eckel-Passow JE, Serie DJ, Bot BM, et al. ANKS1B is a smoking-related molecular alteration in clear cell renal cell carcinoma. BMC Urol 2014;14:14. [Crossref] [PubMed]
- Yusenko MV, Kuiper RP, Boethe T, et al. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer 2009;9:152. [Crossref] [PubMed]
- Wang Z, Jensen MA, Zenklusen JC. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol 2016;1418:111-41. [Crossref] [PubMed]
- Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013;45:860-7. [Crossref] [PubMed]
- Chen F, Chandrashekar DS, Varambally S, et al. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun 2019;10:5679. [Crossref] [PubMed]
- Thul PJ, Åkesson L, Wiking M, et al. A subcellular map of the human proteome. Science 2017;356:eaal3321. [Crossref] [PubMed]
- Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583-9. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021;49:D605-12. [Crossref] [PubMed]
- Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Ray S, Cheaib JG, Biles MJ, et al. Local and Regional Recurrences of Clinically Localized Renal Cell Carcinoma after Nephrectomy: A 15 Year Institutional Experience with Prognostic Features and Oncologic Outcomes. Urology 2021;154:201-7. [Crossref] [PubMed]
- Gorin MA, Patel HD, Rowe SP, et al. Neoadjuvant Nivolumab in Patients with High-risk Nonmetastatic Renal Cell Carcinoma. Eur Urol Oncol 2022;5:113-7. [Crossref] [PubMed]
- Zhang M, Wei T, Zhang X, et al. Targeting lipid metabolism reprogramming of immunocytes in response to the tumor microenvironment stressor: A potential approach for tumor therapy. Front Immunol 2022;13:937406. [Crossref] [PubMed]
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov 2022;12:31-46. [Crossref] [PubMed]
- Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: Still emerging. Cell Metab 2022;34:355-77. [Crossref] [PubMed]
- Sciacovelli M, Dugourd A, Jimenez LV, et al. Dynamic partitioning of branched-chain amino acids-derived nitrogen supports renal cancer progression. Nat Commun 2022;13:7830. [Crossref] [PubMed]
- Wettersten HI, Aboud OA, Lara PN Jr, et al. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol 2017;13:410-9. [Crossref] [PubMed]
- Tan SK, Hougen HY, Merchan JR, et al. Fatty acid metabolism reprogramming in ccRCC: mechanisms and potential targets. Nat Rev Urol 2023;20:48-60. [Crossref] [PubMed]
- Wettersten HI. Reprogramming of Metabolism in Kidney Cancer. Semin Nephrol 2020;40:2-13. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Herman MA, Birnbaum MJ. Molecular aspects of fructose metabolism and metabolic disease. Cell Metab 2021;33:2329-54. [Crossref] [PubMed]
- Xia H, Wang J, Guo X, et al. Identification of a Hypoxia-Related Gene Signature for Predicting Systemic Metastasis in Prostate Cancer. Front Cell Dev Biol 2021;9:696364. [Crossref] [PubMed]
- Lian J, Xia L, Chen Y, et al. Aldolase B impairs DNA mismatch repair and induces apoptosis in colon adenocarcinoma. Pathol Res Pract 2019;215:152597. [Crossref] [PubMed]
- He X, Li M, Yu H, et al. Loss of hepatic aldolase B activates Akt and promotes hepatocellular carcinogenesis by destabilizing the Aldob/Akt/PP2A protein complex. PLoS Biol 2020;18:e3000803. [Crossref] [PubMed]
- Caspi M, Perry G, Skalka N, et al. Aldolase positively regulates of the canonical Wnt signaling pathway. Mol Cancer 2014;13:164. [Crossref] [PubMed]
- Bu P, Chen KY, Xiang K, et al. Aldolase B-Mediated Fructose Metabolism Drives Metabolic Reprogramming of Colon Cancer Liver Metastasis. Cell Metab 2018;27:1249-1262.e4. [Crossref] [PubMed]
- Li M, He X, Guo W, et al. Aldolase B suppresses hepatocellular carcinogenesis by inhibiting G6PD and pentose phosphate pathways. Nat Cancer 2020;1:735-47. [Crossref] [PubMed]
- Wang J, Wu Q, Qiu J. Accumulation of fructose 1,6-bisphosphate protects clear cell renal cell carcinoma from oxidative stress. Lab Invest 2019;99:898-908. [Crossref] [PubMed]
- Nouhaud FX, Blanchard F, Sesboue R, et al. Clinical Relevance of Gene Copy Number Variation in Metastatic Clear Cell Renal Cell Carcinoma. Clin Genitourin Cancer 2018;16:e795-805. [Crossref] [PubMed]
- Capogrosso P, Larcher A, Sjoberg DD, et al. Risk Based Surveillance after Surgical Treatment of Renal Cell Carcinoma. J Urol 2018;200:61-7. [Crossref] [PubMed]
- Parker WP, Cheville JC, Frank I, et al. Application of the Stage, Size, Grade, and Necrosis (SSIGN) Score for Clear Cell Renal Cell Carcinoma in Contemporary Patients. Eur Urol 2017;71:665-73. [Crossref] [PubMed]
- Grimm MO, Leucht K, Foller S. Risk Stratification and Treatment Algorithm of Metastatic Renal Cell Carcinoma. J Clin Med 2021;10:5339. [Crossref] [PubMed]
- Jin X, Wang D, Lei M, et al. TPI1 activates the PI3K/AKT/mTOR signaling pathway to induce breast cancer progression by stabilizing CDCA5. J Transl Med 2022;20:191. [Crossref] [PubMed]
- Li H, Qi Z, Niu Y, et al. FBP1 regulates proliferation, metastasis, and chemoresistance by participating in C-MYC/STAT3 signaling axis in ovarian cancer. Oncogene 2021;40:5938-49. [Crossref] [PubMed]
- Li K, Zhu X, Yuan C. Inhibition of miR-185-3p Confers Erlotinib Resistance Through Upregulation of PFKL/MET in Lung Cancers. Front Cell Dev Biol 2021;9:677860. [Crossref] [PubMed]
- Li H, Wang J, Xu H, et al. Decreased fructose-1,6-bisphosphatase-2 expression promotes glycolysis and growth in gastric cancer cells. Mol Cancer 2013;12:110. [Crossref] [PubMed]
- Shen J, Jin Z, Lv H, et al. PFKP is highly expressed in lung cancer and regulates glucose metabolism. Cell Oncol (Dordr) 2020;43:617-29. [Crossref] [PubMed]
- Hyroššová P, Milošević M, Škoda J, et al. Effects of metabolic cancer therapy on tumor microenvironment. Front Oncol 2022;12:1046630. [Crossref] [PubMed]
- Faraj JA, Al-Athari AJH, Mohie SED, et al. Reprogramming the tumor microenvironment to improve the efficacy of cancer immunotherapies. Med Oncol 2022;39:239. [Crossref] [PubMed]
- Eulberg D, Frömming A, Lapid K, et al. The prospect of tumor microenvironment-modulating therapeutical strategies. Front Oncol 2022;12:1070243. [Crossref] [PubMed]
- Zhang J, Hu Z, Horta CA, et al. Regulation of epithelial-mesenchymal transition by tumor microenvironmental signals and its implication in cancer therapeutics. Semin Cancer Biol 2023;88:46-66. [Crossref] [PubMed]
- Popova NV, Jücker M. The Functional Role of Extracellular Matrix Proteins in Cancer. Cancers (Basel) 2022;14:238. [Crossref] [PubMed]
- Melaiu O, Lucarini V, Cifaldi L, et al. Influence of the Tumor Microenvironment on NK Cell Function in Solid Tumors. Front Immunol 2020;10:3038. [Crossref] [PubMed]
- Jiang X, Liu B, Nie Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 2021;6:74. [Crossref] [PubMed]
- Fang Z, Mei W, Qu C, et al. Role of m6A writers, erasers and readers in cancer. Exp Hematol Oncol 2022;11:45. [Crossref] [PubMed]
- Liu Z, Zou H, Dang Q, et al. Biological and pharmacological roles of m(6)A modifications in cancer drug resistance. Mol Cancer 2022;21:220. [Crossref] [PubMed]
- Deng LJ, Deng WQ, Fan SR, et al. m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol Cancer 2022;21:52. [Crossref] [PubMed]


