
Biomarker identification of immune-related genes in pheochromocytoma and paraganglioma
Highlight box
Key findings
• Presents study found that immunity is closely related to the occurrence of PPGL.
What is known and what is new?
• In recent years, microarray analysis and sequencing technology has become a powerful tool for screening pathogenic genes, and can be used to identify biomarkers with diagnostic and therapeutic value.
• FGF2, FYN, and VCAM1 were firstly reported may ast as the significant genes for PPGL incidence.
What is the implication, and what should change now?
• FGF2, FYN, and VCAM1 can be used as potential diagnosis biomarkers of PPGL, further should collected the PPGL patients samples to perform validated experiments.
Introduction
Pheochromocytoma (PCC) and paraganglioma (PGL), collectively known as PPGL, are rare neuroendocrine tumors mainly secreting catecholamine hormones. PPGL is one of the common causes of secondary hypertension. Data show that the incidence of PPGL is 0.01–0.03%. In general hypertensive outpatients, the rate of PPGL is 0.2–0.6%, and about 5% of patients with adrenal accidental tumors are eventually diagnosed with PPGL (1,2). Studies have shown that about 15–17% of PPGL metastasize, and the treatment options for PPGL after metastasis are limited; the 5-year survival rate is less than 50%, which is the main cause of death (3,4). Current study has shown that it is difficult to determine whether PPGL is metastatic by preoperative biochemical tests and previous pathological results (5). The pathogenesis of PPGL remains unclear, but studies have shown that PPGL has a strong genetic background. More than 20 PPGL susceptibility genes have been reported, and about 50% of patients have germline or system gene mutations (6,7). The adrenal pheochromocytoma score (PASS) and adrenal pheochromocytoma and paraganglioma grading system (GAPP) are commonly used to evaluate the malignant biological characteristics of PPGL (8). However, due to the differences between individuals and the heterogeneity of PPGL, there are also great differences in the changes of pathogenic genes. Depending on the score results and the prognostic information of patients, it can be impossible to accurately evaluate the disease progression, and it is also impossible to conduct in-depth research on the pathogenic causes and find more effective intervention measures. In addition, PPGL is a neuroendocrine disease that secretes a large amount of catecholamines, and whether its pathogenesis involves immunity is still unclear.
In recent years, microarray analysis and sequencing technology has become a powerful tool for screening pathogenic genes, and can be used to identify biomarkers with diagnostic and therapeutic value. Based on bioinformatics, it has been found that KCNQ1 and SCN2A are abnormally methylated genes in PPGL (9). Combined liquid chromatography-tandem mass spectrometry (LC/MS-MS) analysis revealed that COX4I2 and PLAT proteins were highly correlated with PPGL blood supply (10). The Gene Expression Omnibus (GEO) databases provide a wealth of microarray and next-generation sequencing data for human diseases. Exploring specific differentially expressed genes (DEGs) associated with the pathogenesis of PPGL is helpful to better understand the development of PPGL. There was study that have explored the gene aberration of PPGL, and the results showed that PPGL with the low programmed death ligand 1 (PD-L1), without microsatellite instability (11). According to genomics analysis for PPGL, there are several common mutated genes such as FGFR1, NF1, PTEN and so on, but in this study do not have explore the relation between genes mutations and immune microenvironment. In the same year, the study of Chen et al. revealed association between regulated immune microenvironment genes and PGGL progression (12). Here, in this study based on public databases, core DEGs were screened by bioinformatics analysis to explore their possible molecular mechanisms and potential therapeutic drugs, in order to provide a new perspective for the prevention and treatment of PPGL. The following article was presented in accordance with the STARD reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-800/rc).
Methods
Data acquisition and processing
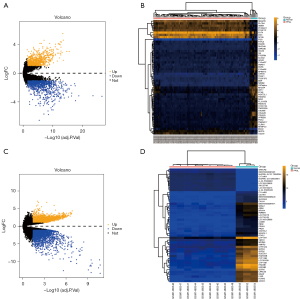
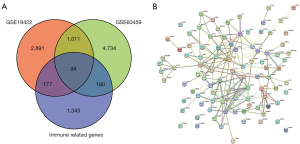
A normal and an adrenal pheochromocytoma cell data set was download from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Finally, GSE19422 (including 84 PPGL and 6 normal adrenal tissue transcriptome datasets) and GSE60459-GPL13607 (including 9 benign PPGL and 3 normal adrenal tissue transcriptome datasets) were included in the subsequent study. Then, the Immunology Database and Analysis Portal Database (ImmPort; https://www.immport.org/home) was used to download the immune-related genes (IRGs), which contained a total of 2,498 IRGs in 17 immunological categories. The “limma” data set was adopted in the R software (The R Foundation of Statistical Computing, Vienna, Austria) for normalization of data, and according to the |logFC| >1 and adjusted P value <0.05. Then, the “ggplot2” package was used to construct the volcano map of the DEGs, and the “pheatmap” package was used to visualize the top 50 DEGs. Finally, the “VennDiagram” package was used to pair the dataset and overlap the IRGs that were considered the IRGs of PPGL. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Protein-protein interaction (PPI) network construction and search for core genes
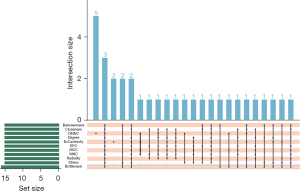
The Search Tool was used for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) to construct a protein-protein interaction (PPI) network for IRGs of PPGL. The tsv format file adapted to Cytoscape (https://cytoscape.org/) was downloaded from the STRING database and the data was poured into Cytoscape. The cytohubba plug-in was then used, which provides analysis algorithms to calculate hub genes in PPI network diagrams. Betweenness, Bottleneck, Closeness, density of maximum neighbourhood component (DMNC), Degree, EcCentricity, edge permeability component (EPC), maximal clique centrality (MCC), maximum neighbourhood component (MNC), Radiality, and Stress were used to calculate the top 15 core genes, and finally, the “UpSetR” package in R language was used to find overlapping genes obtained by 11 algorithms as core genes.
Diagnostic efficiency of core genes and DEGs enrichment analysis
After obtaining the core genes related to PPGL immunity, the author used the “rms” package to construct the receiver operating characteristic (ROC) curve. The area under the curve (AUC) was considered the efficacy of the core genes in diagnosing PPGL, and the 95% confidence interval (CI) was obtained. Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis are databases of gene-related functions stored based on different classifications. GO and KEGG functional enrichment analysis of PPGL immune-related core genes using the “ClusterProfiler” package was performed and the results using circle charts was visualized.
Immune infiltration analysis
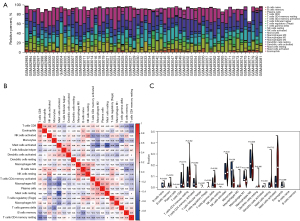
R language was used to analyze the proportion of immune cells between PPGL and normal tissues, and set up 100 repeated operations to calculate the proportion of 22 kinds of immune cells. The normalized expression matrix of GSE19422 dataset was imported into the analysis data, and a P value 0.05 was considered statistically significant. The “ggplot2” in R language was used to construct bar charts to evaluate the proportion of immune cells in each sample, and correlation charts were constructed to evaluate the correlation between immune cells through correlation analysis. Finally, whether there was a difference in the proportion of immune cells between PPGL and normal tissues was compared.
Potential therapeutic drug prediction
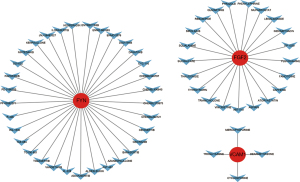
The Drug-Gene Interaction Database (DGIdb; www.dgidb.org) is a drug-gene interaction database that provides information about the association of genes with their known or potential drugs. The core genes related to PPGL immunity were imported into the database, and the therapeutic drugs related to the core genes related to PPGL immunity were obtained and visualized by Cytoscape.
Statistical analysis
R software (version 3.6.3) was used for statistical analysis of all experimental data. The gene expression levels of samples were compared by Student’s t-test, and when P <0.05, the difference was considered statistically significant, and the difference threshold was set as 2 times. In GO and KEGG, q<0.05 was considered statistically significant, an AUC of 0.6–0.7 was considered low performance, 0.7–0.8 was considered medium performance, and >0.8 was considered high performance.
Results
Identification of immune-related DEGs
By differential expression analysis, 963 significantly up-regulated and 1,118 significantly down-regulated genes were identified from the GSE19422 dataset. Totals of 3,219 significantly up-regulated and 2,800 significantly down-regulated genes were identified from GSE60459. The intersection of GSE19422 and GSE60459 data sets and immune genes was used to finally identify 94 immune-related DEGs (Figures 1,2).
PPI analysis and key gene screening
The STRING database was used to construct a PPI network for IRGs of PPGL. The Cytoscape cytoHubba plugin passes the Betweenness, Bottleneck, Closeness, DMNC, Degree, EcCentricity, EPC, MCC, MNC, Radiality, and Stress algorithms, and identified the top 15 genes, and then the common genes in these 11 algorithms were defined as the key genes [fibroblast growth factor 2 (FGF2), FYN proto-oncogene (FYN), and vascular cell adhesion molecule 1 (VCAM1)] (Figures 3,4).

ROC curve analysis
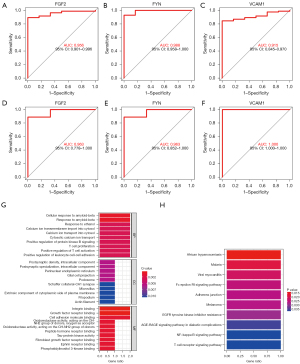
The FGF2, FYN, and VCAM1 genes were analyzed by ROC curve to verify the diagnostic effectiveness of PPGL. The greater the AUC value, the stronger the efficacy of the biomarker in the diagnosis of PPGL, with better specificity and sensitivity. The results showed that FGF2, FYN, and VCAM1 had good diagnostic performance in the GSE19422 and GSE60459 datasets (Figure 5).
Immune-related DEGs enrichment analysis
The GO analysis showed that biological processes were mainly concentrated in cellular response to amyloid-beta and response to ethanol. Cellular component mainly involved postsynaptic density and intracellular component, postsynaptic specialization, intracellular component and perinuclear endoplasmic reticulum. Integrin binding, growth factor receptor binding, and cell adhesion molecule binding were closely related to molecular function. The KEGG enrichment pathway showed that it was mainly related to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance and the AGE-RAGE signaling pathway in diabetic complications; the nuclear factor kappa-B (NF-κB) signaling pathway was correlated with T cell receptor signaling pathway (Figure 5G,5H).
Immune infiltration analysis
To explore the level of immune cell infiltration in PPGL patients, the abundance analysis of immune cell infiltration was performed. The results showed that there were different levels of immune cell infiltration in patients with PPGL. Mast cell activated was significantly increased in patients with PPGL, and was negatively correlated with macrophages M2, mast cell resting, and macrophages M1, and that they were positively correlated with the level of dendritic cells activated (Figure 6).
Drug sensitivity analysis
To explore potential therapeutic agents related to immunity, drug sensitivity prediction was performed. The results showed that more than 50 potential therapeutic agents such as troglitazone, triamcinolone, and Dasatinib were predicted based on the FGF2, FYN, and VCAM1 genes (Figure 7).
Discussion
Studying the difference between diseased tissue and normal tissue is one of the avenues of disease research. PPGL is a rare endocrine tumor. Due to the relatively small amount of clinical data and tissue samples, researchers have limited the exploration of PPGL to a certain extent. In this study, the author hoped to discover genes and therapeutic targets with potential research value by mining the microarray chip of PPGL. By taking the intersection of the chip data, 1,105 DEGs were found, among which 94 were IRGs, accounting for about 10% of DEGs. These results indicate that the formation of PPGL multiple gene mutations and is related to immunity.
3 immune-related DEGs, FGF2, FYN, and VCAM1 were identified, in this study. FGF2, also known as basic fibroblast growth factor (bFGF) (13), is expressed in almost all tissues in the human body. The FGF superfamily mainly includes the paracrine subfamily, endocrine subfamily, intracellular subfamily, and FGF receptor. FGF2 is a member of the paracrine subfamily which is mainly involved in the regulation of proliferation and differentiation of various cells, and it promotes cell migration, division, and proliferation in autocrine or paracrine ways when cells are damaged (14,15). In general, FGF2 functions by binding to FGFR-1c, FGFR-2C, FGFR-3C, and FGFR-1B to form FGF-heparin-FGFR terpolymer complex (16). FGFR/the FGFR signaling pathway is involved in growth and development, wound healing, fibrosis, inflammation, and neovascular formation (17). Study has shown that FGF2 is involved in the regulation of the growth response of the compensatory adrenal gland after unilateral adrenalectomy in rats (18). In addition, because FGF2 has mitogenic effects on cells in the adrenal cortex, it is likely to be a mediator of the N-terminal peptide of adrenocorticotropin (nPOMC) action involved in the regulation of adrenal growth stimulation (19). FYN is a member of the Src kinase family, a non-receptor tyrosine kinase family. It plays a key role in regulating cell proliferation, differentiation, and other biological functions and signal transduction cascades (20). At present, most studies on FYN focus on tumors, and FYN is involved in the progression and metastasis of pancreatic cancer, glioblastoma, and hepatocellular carcinoma (21-23). Dysfunction of FYN is associated with neurodegenerative diseases such as Parkinson’s disease, Alzheimer’s disease, and multiple sclerosis (24). VCAM1 is a surface glycoprotein of endothelial cells. Proinflammatory cytokines and Toll-like receptor agonists, among others, can promote VCAM1 expression (25-27). VCAM1 is expressed by macrophages, dendritic cells, and tumor cells under certain disease conditions with high inflammatory levels (28,29). VCAM1 has been reported to be closely associated with the progression of immune diseases and tumors, and is also a biomarker for predicting cardiovascular diseases (30). At present, there is no relevant report on FYN and VCAM1 in PPGL, which suggests that FYN and VCAM1 may steer the research direction of PPGL in the future.
Enrichment analysis of differentially expressed IRGs showed that PPGL involved multiple pathways. AGE initiates phosphorylation of ERKl/2 and MAPKs by interacting with cell-expressed RAGE receptors, which subsequently leads to abnormal activation of NF-κB, a major regulatory transcription factor of inflammation, which is involved in the expression of inflammatory cytokines and is associated with the occurrence of diabetes-related complications and a variety of chronic diseases. These include cardiovascular diseases, neurodegenerative diseases, arthritis, tumors, and their complications (31-33). Inflammation and oxidative stress are closely related in the pathogenesis of various chronic diseases, and the RAGE/MAPK/NF-KB signaling pathway is the main signaling pathway regulating inflammation and oxidative stress (34). NF-κB has been found to be a nuclear B factor binding to immunoglobulin κ site, which is a conserved inducible transcription factor family. It plays a key role in regulating immune responses, cellular homeostasis, and aging (35,36). In the process of atherosclerosis, the NF-κB pathway is one of the key signaling pathways. After activation, NF-κB can mediate the transcription of various genes, and not only release inflammatory cytokines, but also regulate cell proliferation, apoptosis, morphogenesis and differentiation (37). Blocking the NF-κB pathway can induce apoptosis in chromaffin cells, and NF-κB inhibitors can treat PPGL (38,39). In addition, T cell receptor (TCR) interaction with MHC-antigenic peptide complexes leads to molecular and cellular level changes in T cells, and TCR-mediated T cell activation is also closely related to the NF-κB pathway (40,41). In addition, a significant increase in mast cell activation in PPGL patients was found. These findings suggest that the pathogenesis of PPGL is not only related to genetics, but also closely related to immunity. It is regrettable is that did not have any study investigate these immune related genes roles in PPGL. There was only a study used bioinformatics to explored the immune related genes role in prognosis of PPGL, and the results indicated that ADGRE1, CCL18, and LILRA6 have significant influence on prognosis of PPGL (12). So it is urgent need to further exploration the immune system in PPGL.
Currently, surgery is the preferred treatment for PPGL, regardless of benign and malignant pathological results, timely removal of the tumor is emphasized. Extrusion of the tumor increases the risk of the release of huge quantities of catecholamine, which can increase the risk of acute or chronic complications, so the risk of anesthesia and surgery is higher (42,43). For malignant PPGL or unresectable PPGL, chemotherapy, isotope therapy, and molecular targeted drugs are the main treatment methods. Cyclophosphamide, vincristine, and dacarbazine (CVD), and etoposide and cisplatin (EP) are commonly used chemotherapy regiments that are controlled or effective in about half of PPGL patients (44,45). PPGL has been studied at the molecular level, and the understanding of molecular signaling, metabolism, and resistance mechanisms suggests that treatment regimens can be optimized for each molecular subtype to improve selectivity and effectiveness (46,47). In present study, the author predicted more than 50 drugs based on the specific target genes identified. Among them, troglitazone is a drug used to treat diabetes, and research has shown that it can inhibit the expression of VCAM1 (48). Triamcinolone can inhibit the expression of FGF2, and dasatinib can effectively inhibit FYN (49,50). This study provides a theoretical basis for the development of PPGL-targeted drugs. A shortcoming of this study is that not enough PPGL pathological tissues were collected for verification.
Conclusions
In summary, 94 immune-related DEGs screened in this study are involved in the occurrence and development of PPGL, among which FGF2, FYN, and VCAM1 are key genes, which may be potential biomarkers and therapeutic targets of PPGL.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The author has completed the STARD reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-800/rc
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-800/coif). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lenders JW, Duh QY, Eisenhofer G, et al. Pheochromocytoma and paraganglioma: an endocrine society clinical practice guideline. J Clin Endocrinol Metab 2014;99:1915-42. [Crossref] [PubMed]
- Muth A, Crona J, Gimm O, et al. Genetic testing and surveillance guidelines in hereditary pheochromocytoma and paraganglioma. J Intern Med 2019;285:187-204. [PubMed]
- Corssmit EPM, Snel M, Kapiteijn E. Malignant pheochromocytoma and paraganglioma: management options. Curr Opin Oncol 2020;32:20-6. [Crossref] [PubMed]
- Crona J, Taïeb D, Pacak K. New Perspectives on Pheochromocytoma and Paraganglioma: Toward a Molecular Classification. Endocr Rev 2017;38:489-515. [Crossref] [PubMed]
- Tanabe A, Naruse M. Recent advances in the management of pheochromocytoma and paraganglioma. Hypertens Res 2020;43:1141-51. [Crossref] [PubMed]
- Buffet A, Burnichon N, Favier J, et al. An overview of 20 years of genetic studies in pheochromocytoma and paraganglioma. Best Pract Res Clin Endocrinol Metab 2020;34:101416. [Crossref] [PubMed]
- Li M, Prodanov T, Meuter L, et al. Recurrent disease in patients with sporadic pheochromocytoma and paraganglioma. J Clin Endocrinol Metab 2022;dgac563. Epub ahead of print. [Crossref] [PubMed]
- Neumann HPH, Young WF Jr, Eng C. Pheochromocytoma and Paraganglioma. N Engl J Med 2019;381:552-65. [Crossref] [PubMed]
- Lin D, Lin J, Li X, et al. The Identification of Differentially Expressed Genes Showing Aberrant Methylation Patterns in Pheochromocytoma by Integrated Bioinformatics Analysis. Front Genet 2019;10:1181. [Crossref] [PubMed]
- Sun F, Zhuo R, Ma W, et al. From clinic to mechanism: Proteomics-based assessment of angiogenesis in adrenal pheochromocytoma. J Cell Physiol 2019;234:22057-70. [Crossref] [PubMed]
- Bratslavsky G, Sokol ES, Daneshvar M, et al. Clinically Advanced Pheochromocytomas and Paragangliomas: A Comprehensive Genomic Profiling Study. Cancers (Basel) 2021;13:3312. [Crossref] [PubMed]
- Chen CX, Chen DN, Sun XL, et al. Identification of vital prognostic genes related to tumor microenvironment in pheochromocytoma and paraganglioma based on weighted gene co-expression network analysis. Aging (Albany NY) 2021;13:9976-90. [Crossref] [PubMed]
- D'Amore PA. Modes of FGF release in vivo and in vitro. Cancer Metastasis Rev 1990;9:227-38. [Crossref] [PubMed]
- Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol 2015;4:215-66. [Crossref] [PubMed]
- Dai S, Zhou Z, Chen Z, et al. Fibroblast Growth Factor Receptors (FGFRs): Structures and Small Molecule Inhibitors. Cells 2019.
- Chae YK, Ranganath K, Hammerman PS, et al. Inhibition of the fibroblast growth factor receptor (FGFR) pathway: the current landscape and barriers to clinical application. Oncotarget 2017;8:16052-74. [Crossref] [PubMed]
- Dianat-Moghadam H, Teimoori-Toolabi L. Implications of Fibroblast Growth Factors (FGFs) in Cancer: From Prognostic to Therapeutic Applications. Curr Drug Targets 2019;20:852-70. [Crossref] [PubMed]
- Basile DP, Holzwarth MA. Basic fibroblast growth factor may mediate proliferation in the compensatory adrenal growth response. Am J Physiol 1993;265:R1253-61. [PubMed]
- Fassnacht M, Hahner S, Hansen IA, et al. N-terminal proopiomelanocortin acts as a mitogen in adrenocortical tumor cells and decreases adrenal steroidogenesis. J Clin Endocrinol Metab 2003;88:2171-9. [Crossref] [PubMed]
- Espada J, Martín-Pérez J. An Update on Src Family of Nonreceptor Tyrosine Kinases Biology. Int Rev Cell Mol Biol 2017;331:83-122. [Crossref] [PubMed]
- Gujral TS, Chan M, Peshkin L, et al. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 2014;159:844-56. [Crossref] [PubMed]
- Jiang P, Li Z, Tian F, et al. Fyn/heterogeneous nuclear ribonucleoprotein E1 signaling regulates pancreatic cancer metastasis by affecting the alternative splicing of integrin β1. Int J Oncol 2017;51:169-83. [Crossref] [PubMed]
- Zhang S, Qi Q, Chan CB, et al. Fyn-phosphorylated PIKE-A binds and inhibits AMPK signaling, blocking its tumor suppressive activity. Cell Death Differ 2016;23:52-63. [Crossref] [PubMed]
- Guglietti B, Sivasankar S, Mustafa S, et al. Fyn Kinase Activity and Its Role in Neurodegenerative Disease Pathology: a Potential Universal Target? Mol Neurobiol 2021;58:5986-6005. [Crossref] [PubMed]
- Cook-Mills JM, Marchese ME, Abdala-Valencia H. Vascular cell adhesion molecule-1 expression and signaling during disease: regulation by reactive oxygen species and antioxidants. Antioxid Redox Signal 2011;15:1607-38. [Crossref] [PubMed]
- Osborn L, Hession C, Tizard R, et al. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell 1989;59:1203-11. [Crossref] [PubMed]
- Rice GE, Bevilacqua MP. An inducible endothelial cell surface glycoprotein mediates melanoma adhesion. Science 1989;246:1303-6. [Crossref] [PubMed]
- Sharma R, Sharma R, Khaket TP, et al. Breast cancer metastasis: Putative therapeutic role of vascular cell adhesion molecule-1. Cell Oncol (Dordr) 2017;40:199-208. [Crossref] [PubMed]
- van Oosten M, van de Bilt E, de Vries HE, et al. Vascular adhesion molecule-1 and intercellular adhesion molecule-1 expression on rat liver cells after lipopolysaccharide administration in vivo. Hepatology 1995;22:1538-46. [Crossref] [PubMed]
- Troncoso MF, Ortiz-Quintero J, Garrido-Moreno V, et al. VCAM-1 as a predictor biomarker in cardiovascular disease. Biochim Biophys Acta Mol Basis Dis 2021;1867:166170. [Crossref] [PubMed]
- Henle T. Protein-bound advanced glycation endproducts (AGEs) as bioactive amino acid derivatives in foods. Amino Acids 2005;29:313-22. [Crossref] [PubMed]
- Kellow NJ, Coughlan MT. Effect of diet-derived advanced glycation end products on inflammation. Nutr Rev 2015;73:737-59. [Crossref] [PubMed]
- Zhou Q, Gong J, Wang M. Phloretin and its methylglyoxal adduct: Implications against advanced glycation end products-induced inflammation in endothelial cells. Food Chem Toxicol 2019;129:291-300. [Crossref] [PubMed]
- Yu W, Tao M, Zhao Y, et al. 4'-Methoxyresveratrol Alleviated AGE-Induced Inflammation via RAGE-Mediated NF-κB and NLRP3 Inflammasome Pathway. Molecules 2018;23:1447. [Crossref] [PubMed]
- Mulero MC, Huxford T, Ghosh G. NF-κB, IκB, and IKK: Integral Components of Immune System Signaling. Adv Exp Med Biol 2019;1172:207-26. [Crossref] [PubMed]
- Williams LM, Gilmore TD. Looking Down on NF-κB. Mol Cell Biol 2020;40:e00104-20. [Crossref] [PubMed]
- Gimbrone MA Jr, García-Cardeña G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res 2016;118:620-36. [Crossref] [PubMed]
- Hu HJ, Zhou SH, Liu QM. Treatment of pheochromocytoma blockade of MAPK pathway inhibition in the NF-κB pathway and bFGF - effect of statins on pheochromocytoma patients. Int J Cardiol 2015;182:161-2. [Crossref] [PubMed]
- Pacak K, Sirova M, Giubellino A, et al. NF-κB inhibition significantly upregulates the norepinephrine transporter system, causes apoptosis in pheochromocytoma cell lines and prevents metastasis in an animal model. Int J Cancer 2012;131:2445-55. [Crossref] [PubMed]
- Matsumoto M, Yamada T, Yoshinaga SK, et al. Essential role of NF-kappa B-inducing kinase in T cell activation through the TCR/CD3 pathway. J Immunol 2002;169:1151-8. [Crossref] [PubMed]
- Shah K, Al-Haidari A, Sun J, et al. T cell receptor (TCR) signaling in health and disease. Signal Transduct Target Ther 2021;6:412. [Crossref] [PubMed]
- Castellani MR, Aktolun C, Buzzoni R, et al. Iodine-131 metaiodobenzylguanidine (I-131 MIBG) diagnosis and therapy of pheochromocytoma and paraganglioma: current problems, critical issues and presentation of a sample case. Q J Nucl Med Mol Imaging 2013;57:146-52. [PubMed]
- Waguespack SG, Rich T, Grubbs E, et al. A current review of the etiology, diagnosis, and treatment of pediatric pheochromocytoma and paraganglioma. J Clin Endocrinol Metab 2010;95:2023-37. [Crossref] [PubMed]
- Niemeijer ND, Alblas G, van Hulsteijn LT, et al. Chemotherapy with cyclophosphamide, vincristine and dacarbazine for malignant paraganglioma and pheochromocytoma: systematic review and meta-analysis. Clin Endocrinol (Oxf) 2014;81:642-51. [Crossref] [PubMed]
- Wang P, Li T, Cui Y, et al. 18F-MFBG PET/CT Is an Effective Alternative of 68Ga-DOTATATE PET/CT in the Evaluation of Metastatic Pheochromocytoma and Paraganglioma. Clin Nucl Med 2022; Epub ahead of print. [Crossref]
- Turchini J, Cheung VKY, Tischler AS, et al. Pathology and genetics of phaeochromocytoma and paraganglioma. Histopathology 2018;72:97-105. [Crossref] [PubMed]
- Zethoven M, Martelotto L, Pattison A, et al. Single-nuclei and bulk-tissue gene-expression analysis of pheochromocytoma and paraganglioma links disease subtypes with tumor microenvironment. Nat Commun 2022;13:6262. [Crossref] [PubMed]
- Cominacini L, Garbin U, Pasini AF, et al. The expression of adhesion molecules on endothelial cells is inhibited by troglitazone through its antioxidant activity. Cell Adhes Commun 1999;7:223-31. [Crossref] [PubMed]
- Spandau UH, Sauder G, Schubert U, et al. Effect of triamcinolone acetonide on proliferation of retinal endothelial cells in vitro and in vivo. Br J Ophthalmol 2005;89:745-7. [Crossref] [PubMed]
- Lindauer M, Hochhaus A. Dasatinib. Recent Results Cancer Res 2010;184:83-102. [Crossref] [PubMed]
(English Language Editor: J. Jones)


