
High PRAS40 mRNA expression and its role in prognosis of clear cell renal cell carcinoma
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common type of kidney malignancy tumor, representing about 80% of all primary malignant kidney tumors (1-5). In most cases, ccRCC is resistant to radiation, cytotoxic and hormone therapies, and surgery is the main treatment for ccRCC (5,6). 60% ccRCC patients die in the first 2–3 years, and 30% of ccRCC patients diagnosed with metastases (2). Therefore, searching for new sensitive biomarkers are important for early diagnosis of ccRCC.
PRAS40 also known as AKT1S1 (AKT serine/threonine kinase 1 substrate 1) was initially identified as a 14-3-3 binding protein (7). PRAS40 has two proline-enriched stretches at the N-terminal region (8). PRAS40 dynamically shifts between the nucleus and the cytoplasm. In the nucleus, PRAS40 forms a nuclear-specific complex with PRL11 (ribosomal protein L11) and contributes to senescence and radio resistance (9). In the cytoplasm, PRAS40 is an important interacting partner of mTORC1 (the mechanistic target of rapamycin complex 1) and negatively regulates mTORC1 signaling (10,11). PRAS40 is a substrate of Akt and mediates the PI3K (phosphatidylinositol 3-kinase)/Akt signaling pathway (12). PRAS40 has many phosphorylation sites, and its function was determined by its phosphorylation (13,14). PRAS40 was phosphorylated at T246 by Akt and at S183 by mTORC1 (15). It has been reported that PRAS40 was upregulated in many tumors, including gastric cancer, prostate cancer, etc. (16,17). However, the correlations between clinical characteristics and PRAS40 mRNA expression of ccRCC patients remains to be investigated.
In this study, we compared the PRAS40 mRNA expression between ccRCC tissues and normal tissues. We evaluated the correlations between clinical characteristics and PRAS40 mRNA expression of ccRCC patients. We investigated the biological pathways that related to PRAS40 using GSEA analysis. Our results suggest that PRAS40 was a promising diagnostic and prognostic biomarker for ccRCC.
Methods
Gene expression and clinical characteristics in TCGA
The relevant data provided by TCGA are publicly available and open-ended, and do not require the approval of the local ethics committee. TCGA_KIRC HiSeqV2_PANCAN data were obtained from TCGA database (https://tcga.xenahubs.net). The levels of PRAS40 mRNA, clinicopathological details and general information of ccRCC were collected.
GSEA
In this study, we analyzed the correlations between PRAS40 mRNA expression and all genes by R (v.3.6.1), then performed GSEA analysis using the cluster Profiler package in R. |ES|>1, P<0.05, and FDR <0.25 were considered to be statistically significant.
Statistical analysis
All statistical analysis was conducted by R. ROC curve was generated using the pROC package. We analyzed the correlations between clinical characteristics and PRAS40 mRNA expression using chi-square test. Kaplan-Meier analysis and Multivariate cox analysis was used to analyzed the prognostic value of PRAS40 mRNA expression. P<0.05 was considered statistically significant.
Results
Clinical characteristics of the ccRCC patients
Clinical and gene expression data of 533 primary tumors and 73 normal samples were download from TCGA database, including patients’ age, gender, grade, overall survival, stage, distant metastasis, survival status, T classification and lymph nodes (Table 1).
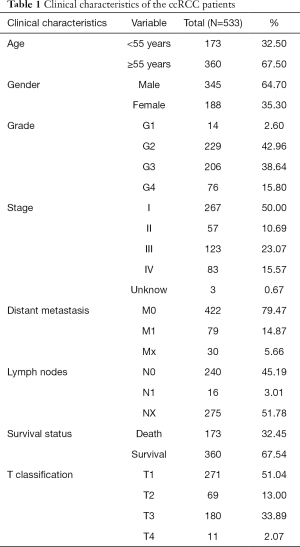
Full table
High PRAS40 expression in ccRCC patients
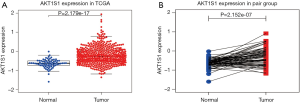
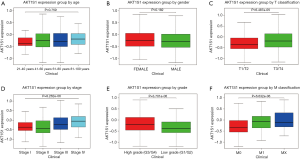
We analyzed the transcription levels of PRAS40 based on TCGA database. We found that PRAS40 mRNA expression was significantly higher in ccRCC tissues than that in normal tissues (P=2.179e-17, P=2.152e-07, respectively) (Figure 1). Furthermore, different PRAS40 mRNA expression were observed in groups based on T classification, clinical stage, distant metastasis and histological grade. The PRAS40 mRNA expression of patients with T3/T4 classification were higher than that of patients with T1/T2 classification (P=6.139e-04). Patients who were in high histological stage had higher PRAS40 mRNA expression than patients who were in low histological stage (P=8.262e-06). Patients with a positive distant metastasis had higher PRAS40 mRNA expression than patients with a negative status. High grade groups (G3/G4) had higher PRAS40 mRNA expression than low grade groups (G1/G2) (P=1.86e-06) (Figure 2).
Diagnostic value of PRAS40 mRNA expression in ccRCC
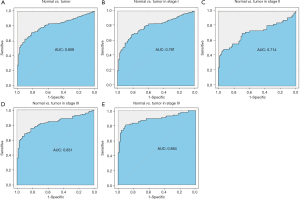
In this study, we evaluated the diagnostic value of PRAS40 mRNA expression by ROC curve. The results showed that the Area Under the Curve (AUC) of PRAS40 was 0.808. We also analyzed the diagnostic capability of PRAS40 mRNA expression in different stages, and the results showed similar diagnostic value with AUC values of 0.791, 0.714, 0.831 and 0.884 for stage I, II, III and IV, respectively (Figure 3).
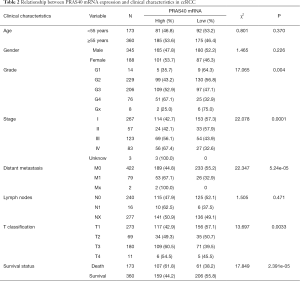
Correlation between clinical characteristics and PRAS40 mRNA expression of ccRCC
In this study, correlations between clinical characteristics and PRAS40 mRNA expression in ccRCC were analyzed by chi-square test. Based on medium PRAS40 mRNA expression, we divided into high or low group. Our results showed that high PRAS40 mRNA expression was associated with T classification (P=0.0033), survival status (P=2.391e-05), histological grade (P=0.004), distant metastasis (P=5.24e-05) and clinical stage (P=0.0001), respectively (Table 2).
Full table
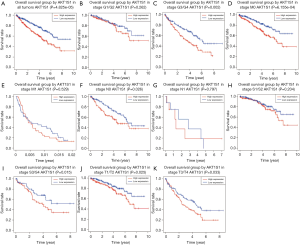
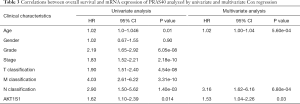
High PRAS40 mRNA expression is an independent risk factor for overall survival in ccRCC patients
Kaplan-Meier analysis revealed that higher PRAS40 mRNA expression was correlated with poor overall survival (P=4.825e-05). Subgroup analysis indicated that PRAS40 mRNA overexpression was significantly affected the overall survival in ccRCC cases of M0 (P=6.155e-04), histological grade G3/G4 (P=0.002), N0 (P=0.026), T1/T2 (P=0.025), T3/T4 (P=0.033) and clinical stage III/IV (P=0.015), respectively (Figure 4). The univariate analysis revealed that higher PRAS40 mRNA expression, advanced stage, high grade, distant metastasis and positive lymph nodes were correlated with overall survival (Table 3). Multivariate analysis showed that PRAS40 mRNA expression was an independent risk factor for overall survival in ccRCC (Table 3).
Full table
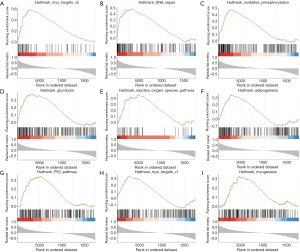
GSEA identifies a PRAS40-related signaling pathway
GSEA was performed to identify the signaling pathways that activated in ccRCC. The results showed that myc targets, DNA repair, oxidative phosphorylation, glycolysis, adipogenesis, p53 pathway, reactive oxygen species pathway and myogenesis were differentially enriched in the positively correlated with PRAS40 mRNA expression phenotype (Table 4, Figure 5).

Full table
Discussion
In this study, our results confirmed that PRAS40 mRNA expression was higher in ccRCC tissues than in normal tissues. High PRAS40 mRNA expression was associated with survival status, distant metastasis, clinical stage, T classification and histological grade, respectively. Moreover, high PRAS40 mRNA expression correlated with poor overall survival. In addition, PRAS40 was an independent prognostic factor of ccRCC.
PRAS40 was aberrantly expressed or hyperphosphorylated in a variety of tumors (16-18). The most common type of PRAS40 alteration is overexpression, PRAS40 was overexpressed in breast cancer, melanoma, colon cancer, etc., but not found in ccRCC yet (19-21). Yuan et al. reported that approximately 40% of trastuzumab resistant HER2-positive patients with p-PRAS40 Thr246 overexpression, and increased risk of tumor progression (22). Ma et al. found that PRAS40 was upregulated and reduction of PRAS40 expression inhibit the proliferation of HCC (23). PRAS40 was overexpressed in Ewing sarcoma family tumors (ESFT), and silence PRAS40 inhibited the proliferation of ESFT cell (24). Consistently, our results revealed that higher PRAS40 mRNA expression in ccRCC. ROC analysis revealed that PRAS40 present a moderate diagnostic value, providing evidence that PRAS40 was a promising biomarker for the diagnosis of ccRCC.
High PRAS40 mRNA expression was significantly correlated with poor prognosis in different tumors (7). Phospho-PRAS40 Thr246 was overexpressed in gastric cancers (17). PRAS40 deletions suppresses the invasion and migration of ESFT and hepatocellular carcinoma (HCC) cells (24,25). Consistently, we found that PRAS40 mRNA expression was significantly associated with poor overall survival, we further investigated the relation between clinical characteristics and PRAS40 mRNA expression in ccRCC patients, we found that histological grade, clinical stage, T classification were highly correlated with PRAS40 mRNA expression. The potential mechanism may involve in myc targets, DNA repair, oxidative phosphorylation, glycolysis, adipogenesis, p53 pathway, reactive oxygen species pathway and myogenesis.
Overall, our study verified the value of PRAS40 mRNA expression in diagnosis and prognosis of ccRCC patients. However, there are some limitations. First, we only conducted bioinformatics mining, our results should be verified in clinical samples. The second limitation is that the current study focuses on the transcription level of PRAS40 but not protein level, the specific mechanism of PRAS40 in ccRCC should be further investigated.
Conclusions
Our results confirmed that PRAS40 mRNA expression was upregulated in ccRCC tissues. Moreover, higher PRAS40 mRNA expression was correlated with poor overall survival of ccRCC patients. In addition, PRAS40 mRNA expression was an independent prognostic factor of ccRCC, making it a promising biomarker with great potential in the future.
Acknowledgments
Funding: This study was supported by the Natural Science Foundation of Fujian Province of China (2019J01508), Youth Science Foundation of Fujian Provincial Health and Family Planning Commission (2017-1-18, 2017-1-19) and Foundations of Fujian Provincial Maternity and Children’s Hospital (YCXZ 19-30), China.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-741
Peer Review File: Available at http://dx.doi.org/10.21037/tau-20-741
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-741). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The relevant data provided by TCGA are publicly available and open-ended, and do not require the approval of the local ethics committee.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Song J, Liu YD, Su J, et al. Systematic analysis of alternative splicing signature unveils prognostic predictor for kidney renal clear cell carcinoma. J Cell Physiol 2019;234:22753-64. [Crossref] [PubMed]
- Mendoza-Alvarez A, Guillen-Guio B, Baez-Ortega A, et al. Whole-Exome Sequencing Identifies Somatic Mutations Associated With Mortality in Metastatic Clear Cell Kidney Carcinoma. Front Genet 2019;10:439-49. [Crossref] [PubMed]
- Qin S, Shi X, Wang C, et al. Transcription Factor and miRNA Interplays Can Manifest the Survival of ccRCC Patients. Cancers (Basel) 2019;11:1668-89. [Crossref] [PubMed]
- Kim B, Jung M, Moon KC. The Prognostic Significance of Protein Expression of CASZ1 in Clear Cell Renal Cell Carcinoma. Dis Markers 2019;2019:1342161. [Crossref] [PubMed]
- Evelonn EA, Landfors M, Haider Z, et al. DNA methylation associates with survival in non-metastatic clear cell renal cell carcinoma. BMC Cancer 2019;19:65-81. [Crossref] [PubMed]
- Yin L, Li W, Wang G, et al. NR1B2 suppress kidney renal clear cell carcinoma (KIRC) progression by regulation of LATS 1/2-YAP signaling. J Exp Clin Cancer Res 2019;38:343-55. [Crossref] [PubMed]
- Lv D, Guo L, Zhang T, et al. PRAS40 signaling in tumor. Oncotarget 2017;8:69076-85. [Crossref] [PubMed]
- Zhang H. MELK is Upregulated in Advanced Clear Cell Renal Cell Carcinoma and Promotes Disease Progression by Phosphorylating PRAS40. Cell Transplant 2019;28:37S-50S. [Crossref] [PubMed]
- Havel JJ, Li Z, Cheng D, et al. Nuclear PRAS40 couples the Akt/mTORC1 signaling axis to the RPL11-HDM2-p53 nucleolar stress response pathway. Oncogene 2015;34:1487-98. [Crossref] [PubMed]
- Wiza C, Nascimento EB, Ouwens DM. Role of PRAS40 in Akt and mTOR signaling in health and disease. Am J Physiol Endocrinol Metab 2012;302:E1453-60. [Crossref] [PubMed]
- Sancak Y, Thoreen CC, Peterson TR, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 2007;25:903-15. [Crossref] [PubMed]
- Zhang KS, Schecker J, Krull A, et al. PRAS40 suppresses atherogenesis through inhibition of mTORC1-dependent pro-inflammatory signaling in endothelial cells. Sci Rep 2019;9:16787. [Crossref] [PubMed]
- Hsu PP, Kang SA, Rameseder J, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011;332:1317-22. [Crossref] [PubMed]
- Wang L, Harris TE, Roth RA, et al. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem 2007;282:20036-44. [Crossref] [PubMed]
- Subbannayya T, Leal-Rojas P, Zhavoronkov A, et al. PIM1 kinase promotes gallbladder cancer cell proliferation via inhibition of proline-rich Akt substrate of 40 kDa (PRAS40). J Cell Commun Signal 2019;13:163-77. [Crossref] [PubMed]
- Shipitsin M, Small C, Giladi E, et al. Automated quantitative multiplex immunofluorescence in situ imaging identifies phospho-S6 and phospho-PRAS40 as predictive protein biomarkers for prostate cancer lethality. Proteome Sci 2014;12:40-53. [Crossref] [PubMed]
- Lu YZ, Deng AM, Li LH, et al. Prognostic role of phospho-PRAS40 (Thr246) expression in gastric cancer. Arch Med Sci 2014;10:149-53. [Crossref] [PubMed]
- Vincent EE, Elder DJ, Thomas EC, et al. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. Br J Cancer 2011;104:1755-61. [Crossref] [PubMed]
- Malla R, Ashby CR Jr, Narayanan NK, et al. Proline-rich AKT substrate of 40-kDa (PRAS40) in the pathophysiology of cancer. Biochem Biophys Res Commun 2015;463:161-6. [Crossref] [PubMed]
- Van Der Steen N, Leonetti A, Keller K, et al. Decrease in phospho-PRAS40 plays a role in the synergy between erlotinib and crizotinib in an EGFR and cMET wild-type squamous non-small cell lung cancer cell line. Biochem Pharmacol 2019;166:128-38. [Crossref] [PubMed]
- Rho O, Srivastava J, Cho J, et al. Overexpression of PRAS40(T246A) in the Proliferative Compartment Suppresses mTORC1 Signaling, Keratinocyte Migration, and Skin Tumor Development. J Invest Dermatol 2016;136:2070-9. [Crossref] [PubMed]
- Yuan K, Wu H, Wang Y, et al. Phospho-PRAS40(Thr246) predicts trastuzumab response in patients with HER2-positive metastatic breast cancer. Oncol Lett 2015;9:785-9. [Crossref] [PubMed]
- Ma X, Yan J, Chen W, et al. Knockdown of Myosin VI Inhibits Proliferation of Hepatocellular Carcinoma Cells In Vitro. Chem Biol Drug Des 2015;86:723-30. [Crossref] [PubMed]
- Huang L, Nakai Y, Kuwahara I, et al. PRAS40 is a functionally critical target for EWS repression in Ewing sarcoma. Cancer Res 2012;72:1260-9. [Crossref] [PubMed]
- Nagaraju GP, Bramhachari PV, Raghu G, et al. Hypoxia inducible factor-1alpha: Its role in colorectal carcinogenesis and metastasis. Cancer Lett 2015;366:11-8. [Crossref] [PubMed]


