
Effect of upregulation of DD3 on early detection and prognosis in prostate cancer
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related death among men in the United States (1). Its incidence has increased at a tremendous rate among Asian male populations in the past decade (2). Most PCa patients are diagnosed at an advanced stage, requiring curative surgery with extended lymphadenectomy. Traditional treatment for PCa primarily consists of radical surgery and radiotherapy, and is only effective for early-stage PCa without metastasis. Presently, no curative therapy is available for PCa with local or distant metastases, resulting in poor prognosis for patients with advanced disease (3). Therefore, there is a pressing need to discover effective diagnostic biomarkers for the detection of early-stage PC. Biopsies, digital rectal examination, and transrectal ultrasound are current techniques for PCa detection, but they perform poorly as early detection tools.
Prostate-specific antigen (PSA) is the most widely used biomarker for malignancy screening in prostate epithelium biopsy analysis and is considered the gold standard for the diagnosis of PCa in clinical practice (4). In the routine initial screening procedures for PCa, a high PSA level (≥4 ng/mL) indicates the requirement of a prostatic puncture biopsy with histopathological analysis for the differential diagnosis of PCa from other prostatic diseases (5). As elevated PSA levels are also observed in patients with benign prostate hyperplasia (BPH) and prostatitis, unnecessary puncture biopsies are often performed. In one study, approximately 15% of men with PSA levels below the cut-off value of 4 ng/mL were reported to be at risk for PCa (6). Additionally, two randomized trials evaluated the effects of PSA-based screening on patient mortality, but produced conflicting results (7,8). These data overwhelmingly reveal that PSA is not a PCa-specific biomarker. Moreover, PSA level does not indicate PCa risk or distinguish indolent from aggressive disease, particularly when the level is below 20 ng/mL. Because PSA is an inadequate tumor biomarker, there is an urgent need to identify novel high-fidelity PCa biomarkers.
Prostate cancer antigen 3 (DD3), a long non-coding RNA also known as differential display code 3, is expressed 10- to 100-fold in PCa tissues compared with adjacent non-cancerous tissues. It locates on the long arm of chromosome 9, containing 3 introns and 4 exons (9). In normal tissues and other prostatic lesion tissues, DD3 levels were found below detection range by Northern blot analysis (9), making it an ideal biomarker for detecting PCa (10). Several studies have reported that DD3 expression can be detected across multiple biological sample types, such as blood and urine, serving as a specific tumor marker for PCa (11). One study showed that DD3 was highly expressed in 95% of PCa tumor cells with a median 66- to 100-fold upregulation compared with adjacent non-neoplastic cells (12).
It is feasible to understand the variable expression of DD3, specifically as it relates to tumorigenesis and the development of PCa. For example, expression of DD3 mRNA was demonstrated to be androgen-regulated via the activation of androgen receptor (AR)-mediated signaling, with the potential to regulate PCa cell survival (13). Another study reported that DD3 silencing resulted in the loss of PCa cell viability with regulatory effects on multiple tumor-related genes, the majority of which were linked to epithelial-mesenchymal transition markers and AR cofactors (14). As a pro-cancer marker, DD3 was observed to play a role in increase of cell proliferation, invasion, migration, and inhibition of apoptosis (15). The wealth of evidence supporting DD3 as a biomarker of PCa led to the development of a commercial kit named Progensa®. This DD3 assay was manufactured by GenProbe and employs a PSA mRNA internal control for the quantification of DD3 in urine samples (16). In urine, several cancer products including DD3 that are released through prostate ducts act as cell-free markers (17). Salagierski et al. (18) also supported DD3 mRNA transcripts as being detectable and quantifiable in urine. However, the detection of DD3 gene expression in urine is prone to error due to the different degrees of prostatic massage among patients and the unhygienic extraction of urine samples. Thus, detecting DD3 in the blood may be a more reliable strategy for biomarker detection and PCa screening.
In this study, the SYBR Green assay was used to detect copies of the DD3 gene in blood. Detection efficacy was then further validated in PCa tissues. For the first time, DD3 expression levels in patients were compared before and after surgical intervention. The Cancer Genome Atlas (TCGA) database, DD3 mRNA expression, and PCa patient mortality data were used to elucidate the role of DD3 in tumorigenesis and progression.
Methods
Patients and clinical specimens
Forty-one human prostate tumor tissue samples and 21 adjacent normal tissues were collected from PCa patients who underwent curative resection of the tumor at Yantai Yuhuangding Hospital (Shandong, China) between 2012 and 2016 immediately after surgery and stored at −80 °C for further use. Tissue sample size was verified by a pathologist and the paired peritumoral samples were confirmed to be free of tumor deposits. Blood samples were collected in BD PAXgene Blood RNA tubes (Qiagen, Hilden, Germany) and stored in aliquots at −80 °C. Peripheral blood samples were obtained from 53 patients with BPH and 102 patients with PCa, and also from 35 PCa patients before and after operation. PCa was confirmed by prostatic biopsy and histopathological evaluation to distinguish benign epithelial cells (in BPH samples) from adenocarcinoma cells (in PCa samples). Each tumor was classified by two pathologists and graded using the Gleason scoring system. All blood and tissue specimens were obtained from consenting subjects. This study strictly followed protocols approved by the Institutional Review Board and Research Ethical Committee of Yantai Yuhuangding Hospital of Qingdao University.
RNA isolation and reverse transcription
Total blood RNA was isolated using the PAXgene Blood RNA Kit (Qiagen, Germany), and total tissue RNA was isolated from PCa specimens using the ABgene Total RNA Isolation Reagent (Advanced Biotechnologies Ltd., Epsom, Surrey, UK). RNA concentration and quality were analyzed with the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, RRID:SCR_018042). Following the manufacturer’s instructions, cDNA was generated from total RNA using the Quantitect Reverse Transcription Kit (Qiagen, Germany). Quantified analysis of DD3 mRNA was conducted using real-time quantitative polymerase chain reaction (RT-qPCR) (Funglyn Biotech Co., Ltd, Shanghai, China) and the QuantiNova SYBR Green PCR Kit (Qiagen, Germany). The primer sequences were as follows: DD3 forward primer (5’-ACACAGGAAGCACAAAAGG-3’) and reverse primer (5’-GATGACCCAAGATGGCGGC-3’), and β-actin forward primer (5’-CTGGACTTCGAGCAAGAGATG-3’) and reverse primer (5’-GAGTTGAAGGTAGTTTCGTGGA-3’). RT-qPCR reaction was conducted at 95 °C for 2 min, 95 °C for 5 s and 60 °C for 10 s for 40 cycles, followed by melt curve analysis. 2(−Δct)×106 was used as the quantitative threshold for gene expression.
TCGA data
Dataset of mRNA levels (RNASeq v2) in PCa patients were downloaded from the TCGA database (www.synapse.org, The Cancer Genome Atlas, RRID:SCR_003193), and the DD3 mRNA levels and whole transcriptome data were used. Clinical data were obtained from the cBioPortal database (www.cbioportal.org, RRID:SCR_014555).
Gene set enrichment analysis
The associations between DD3 mRNA expression level and biological processes and signaling pathways were analyzed using gene set enrichment analysis software (GSEA v2.2, software.broadinstitute.org/gsea/, RRID:SCR_003199). GSEA calculated an estimated gene set enrichment score as a function of a predefined gene set of high- and low-ranked PCa genes distributed randomly (Molecular Signatures Database, software.broadinstitute.org, gene sets: TUMORIGENESIS, PROSTATE_CANCER). Default settings of the software were utilized. Significance was determined by permutation analysis (1,000 permutations) and false discovery rate (FDR). A gene set was considered significantly enriched when the FDR was <0.05.
Statistical analysis
The results from the RT-qPCR of paired samples (i.e., blood from a patient before and after surgery) were analyzed by the paired t-test. Results of RNA-seq v2 and the qPCR of unpaired samples were analyzed by the independent samples t-test. Statistical Package for Social Sciences (version 19.0; SPSS Inc., Stanford, CA, USA, RRID:SCR_002865) and GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA, USA, RRID:SCR_002798) were used for statistical analyses. To assess the sensitivity and specificity of various biomarkers associated with the diagnosis of prostate abnormalities, receiver operating characteristic (ROC) curve analysis was applied using an estimation of the area under the curve (AUC). The log-rank test for the generated Kaplan-Meier curve was performed to evaluate the association between the DD3 mRNA expression level and patient mortality. Overall survival (OS) was defined as the time between the first surgery for primary PCa and death. Disease-free survival was defined as the time between the first surgery and disease recurrence. Logistic regression analysis was conducted to confirm the analysis results. Observations were considered statistically significant when two-tailed P values <0.05. Data values are reported as the mean ± standard error of the mean unless otherwise noted.
Results
Elevated DD3 mRNA expression in PCa tissues
DD3 mRNA contains four exons. We selected an appropriate exon of DD3 mRNA for amplification according to a recent report (19). The expression patterns of DD3 were examined across 62 tissue specimens using the described SYBR Green RT-qPCR assay. A total of 41 PCa tissues and 21 adjacent normal tissues were analyzed. DD3 expression was significantly increased (P=0.013) in PCa tumors compared with paired adjacent prostate tissues (Figure 1A). By analyzing data from the publicly available TCGA database we further validated the significant increase of DD3 expression in PCa tissues (P<0.0001, Figure 1B).
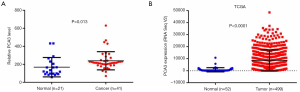
DD3 upregulation associated with poor outcome
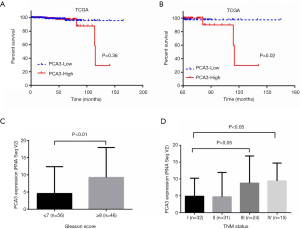
To evaluate the value of DD3 expression level as a prognostic marker of PCa, patients from TCGA dataset were divided into high DD3 expression and low DD3 expression groups by the median level of DD3 The Kaplan-Meier curve suggested a trend linking shorter OS with high DD3 expression than with low DD3 expression (P=0.36, Figure 2A). Further analysis of patients who survived longer than 5 years showed that patients with low DD3 expression had longer OS (P=0.02, Figure 2B). The Gleason score and TNM stage of PCa were important manifestation of disease progression. We analyzed the relationship between DD3 expression and Gleason score and TNM stage of PCa from TCGA dataset. The expression of DD3 in patients with Gleason score ≥8 was higher than in those with Gleason score ≤7 (P<0.01, Figure 2C). Also the expression levels of DD3 in PCa was correlated with TNM status (P<0.05, Figure 2D).
Different blood DD3 mRNA level between PCa and BPH patients
RNA was isolated in vitro and purified from the peripheral blood samples of a total of 155 patients, including 102 (65.81%) and 53 (34.19%) cases of biopsy-confirmed PCa and BPH, respectively (Table 1). The optical density at 260 nm (OD260) over that at 280 nm (OD280) of RNA ranged from 1.8 to 2.0. Patients’ age ranged from 52 to 89 years. The relative mRNA expression of DD3 was significantly lower in BPH patients (2.87±6.88) than in PCa patients (6.77±8.49) (P=0.005). However, there was no significant difference in patient’s age (P=0.12), free PSA (fPSA) levels (P=0.546) or total PSA levels (P=0.659) between patients with BPH and patients with PCa (Table 1).

Full table
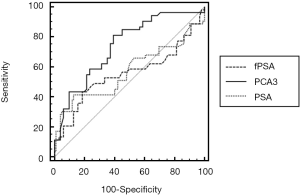
Better diagnostic value for DD3 than total and free PSA
A ROC curve was used to illustrate the diagnostic performances of DD3 mRNA and PSA for the detection of PCa (Table 2). In particular, the diagnostic performance of DD3 mRNA, fPSA, and PSA with regard to sensitivity and specificity was demonstrated (Figure 3). The AUC of both PSA and fPSA was smaller than that of DD3 mRNA. The DD3 mRNA showed a significant correlation with the clinical diagnosis of PCa (P<0.0001).

Full table
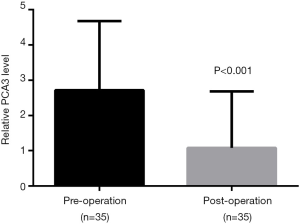
Blood DD3 decreased after prostatectomy
To evaluate prognostic value of blood DD3 mRNA level in PCa, blood specimens from 35 PCa patients were collected before and after the operation. Total RNA from whole blood was isolated and subjected to SYBR Green-based RT-qPCR to measure DD3 expression. The results showed that DD3 expression in the blood was significantly decreased postoperatively compared to preoperative samples (P=0.0016), suggesting that it might act as an effective tumor prognostic marker (Figure 4).
DD3 associated with PCa tumorigenesis and proliferation
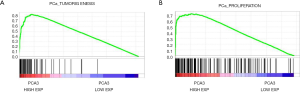
To address the mechanisms of DD3-driven tumorigenesis, TCGA PCa samples were analyzed with GSEA. Intriguingly, GSEA showed significant enrichment of tumorigenesis-related genes in samples with high DD3 expression, suggesting that DD3 might participate in the tumorigenesis of PCa (Figure 5A). Then, GSEA was used to further explore the correlation of DD3 expression level with cell proliferation in PCa patients. PCa patient data from the TCGA dataset were divided into high and low DD3 expression groups by the median level of DD3. Correlation between DD3 expression level and cell proliferation gene set was analyzed (Figure 5B). The gene set correlating with cell proliferation was highly enriched in the DD3 high expression group, suggesting that DD3 expression level was positively correlated with cell proliferation.
Discussion
Currently, serum PSA levels are widely used to screen patients for PCa. According to the European Association of Urology and the National Comprehensive Cancer Network guidelines, a digital rectal examination followed by histological analysis of prostate biopsy is the golden standard for PCa diagnosis (20,21). However, the specificity of PSA detection for PCa is low, and approximately 75% of men who are clinically suspected of PCa have a negative biopsy result (22,23). Thus, it remains a challenge to explore and develop a diagnostic signature of PCa through the application of novel high-fidelity biomarkers. Diagnosis of PCa has improved with the identification of new biomarkers, and data suggested that implementation of DD3 testing is reducing the number of unnecessary biopsies.
DD3 was the first PCa-related non-coding RNA shown to be specifically expressed in prostate tissues and only overexpressed in PCa tissues (24). The clinical application of DD3 has been established as an independent diagnostic biomarker for the prediction of PCa in initial and repeated biopsy environments, primarily through urine-based and biopsy analyses (25). Studies have reported that DD3 expression was high in 95% of PCa tissues compared with paired adjacent tissues (26,27). DD3 has been shown to be a poor marker in the initial biopsy setting with a high false negative rate and low sensitivity across several studies, leading to the underdiagnosis of high-grade disease (28,29). However, DD3 mRNA transcripts can be detected and quantified as a cell-free marker in urine samples (17,18). In addition, urinary DD3 mRNA provides a more accurate PCa diagnostic target than PSA RNA and serum tPSA (19,30,31). Given the limitations inherit to sample acquisition of biopsy tissue and urine, specifically puncture biopsy, and lack of sample collection standardization, the downstream evaluation of DD3 is limited. Unlike urine, blood sample acquisition is clean and standardized, resulting in dramatically improved sample reliability.
In this study, DD3 mRNA expression was significantly higher in PCa tissues than in paired adjacent tissues, as determined by RT-qPCR analysis. The expression of DD3 was verified through analysis of the TCGA database. Ferreira et al. (13) reported DD3 might modulate PCa cell survival. This study examined DD3 as it relates to PCa patient prognosis and survival. The Kaplan-Meier curve suggested that low DD3 expression led to longer OS than did high DD3 expression. ROC curve has been well documented as an effective way to assess the sensitivity and specificity of multiple biomarkers for diagnostic applications. Thus, in this study, ROC curve analysis was used to compare the diagnostic efficacy of DD3 comparing with PSA and fPSA in PCa. The AUC of DD3 mRNA was larger than both PSA and fPSA, suggesting that DD3 is a better tumor biomarker for the effective diagnosis of PCa. Previous data have demonstrated increased sensitivity and specificity when using DD3 copy number as a PCa diagnostic marker compared with PSA copy number, as determined by TaqMan-based RT-qPCR (32). Furthermore, the overall diagnostic efficacy of DD3 RNA is higher than that of PSA RNA (19).
There has been contradiction in the AUC data of DD3 in the literature (33,34), which might be an artifact of their small sample sizes. Following this expanded sample number analysis, the difference of AUC between DD3 and PSA in our study was closer to what had been previously reported in the literature. Interestingly, DD3 expression in the blood was significantly decreased in postoperative samples compared with preoperative samples, further suggesting DD3 as an effective tumor prognostic marker, a finding that has not been previously reported.
The signaling pathway of DD3 in PCa is gradually being elucidated. Pro-survival signals mediated by AR are key factors in the development of PCa due to the known impacts on PCa cell proliferation, differentiation, and survival (35). In addition, DD3 promotes cell proliferation, invasion, and migration and inhibits apoptosis (15). Our GSEA analysis showed that high DD3 expression was correlated with PCa proliferation-associated genes.
Acknowledgments
Funding: This study was funded by the National Natural Science Foundation of the People's Republic of China (Grant No. 81372367 and 81402292), the Yantai Key Research and Development Project (Grant No. 2017YD005), and the Shandong Provincial Natural Science Foundation, China (Grant No. ZR2017PH047).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-19-899
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-19-899). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional research board of ethical committee at the Affiliated Yantai Yuhuangding Hospital of Qingdao University (No.: 2020-168) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pernar CH, Ebot EM, Wilson KM, et al. The epidemiology of prostate cancer. Cold Spring Harb Perspect Med 2018;8:a030361. [Crossref] [PubMed]
- Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol 2017;3:524-48. [Crossref] [PubMed]
- Thoma C. Prostate cancer: Predicting risk of metastasis after prostatectomy. Nat Rev Urol 2018;15:137. [PubMed]
- Graham J, Baker M, Macbeth F, et al. Diagnosis and treatment of prostate cancer: summary of NICE guidance. Bmj 2008;336:610-12. [Crossref] [PubMed]
- Koie T, Mitsuzuka K, Yoneyama T, et al. Prostate-specific antigen density predicts extracapsular extension and increased risk of biochemical recurrence in patients with high-risk prostate cancer who underwent radical prostatectomy. Int J Clin Oncol 2015;20:176-81. [Crossref] [PubMed]
- Thompson IM, Pauler DK, Goodman PJ, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level≤ 4.0 ng per milliliter. New Engl J Med 2004;350:2239-46. [Crossref] [PubMed]
- Andriole GL, Crawford ED, Grubb RL III, et al. Prostate cancer screening in the randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: mortality results after 13 years of follow-up. J Natl Cancer Inst 2012;104:125-32. [Crossref] [PubMed]
- Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate cancer mortality: results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet 2014;384:2027-35. [Crossref] [PubMed]
- Bussemakers MJ, Van Bokhoven A, Verhaegh GW, et al. DD3: A new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999;59:5975-79. [PubMed]
- Hendriks RJ, Van Oort I, Schalken J. Blood-based and urinary prostate cancer biomarkers: a review and comparison of novel biomarkers for detection and treatment decisions. Prostate Cancer Prostatic Dis 2017;20:12. [Crossref] [PubMed]
- Dijkstra S, Mulders P, Schalken J. Clinical use of novel urine and blood based prostate cancer biomarkers: a review. Clin Biochem 2014;47:889-96. [Crossref] [PubMed]
- Herness EA, Naz RK. A novel human prostate-specific gene-1 (HPG-1): molecular cloning, sequencing, and its potential involvement in prostate carcinogenesis. Cancer Res 2003;63:329-36. [PubMed]
- Ferreira LB, Palumbo A, de Mello KD, et al. PCA3 noncoding RNA is involved in the control of prostate-cancer cell survival and modulates androgen receptor signaling. BMC Cancer 2012;12:507. [Crossref] [PubMed]
- Lemos AEG, Ferreira LB, Batoreu NM, et al. PCA3 long noncoding RNA modulates the expression of key cancer-related genes in LNCaP prostate cancer cells. Tumour Biol 2016;37:11339-48. [Crossref] [PubMed]
- Lin C, Wang J, Wang Y, et al. GRP78 Participates in PCA3-regulated prostate cancer progression. Anticancer Res 2017;37:4303-10. [PubMed]
- Groskopf J, Aubin SM, Deras IL, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem 2006;52:1089-95. [Crossref] [PubMed]
- Truong M, Yang B, Jarrard DF. Toward the detection of prostate cancer in urine: a critical analysis. J Urol 2013;189:422-9. [Crossref] [PubMed]
- Salagierski M, Schalken JA. Molecular diagnosis of prostate cancer: PCA3 and TMPRSS2: ERG gene fusion. J Urol 2012;187:795-801. [Crossref] [PubMed]
- Wang T, Qu X, Jiang J, et al. Diagnostic significance of urinary long non-coding PCA3 RNA in prostate cancer. Oncotarget 2017;8:58577. [Crossref] [PubMed]
- Mohler JL. The 2010 NCCN clinical practice guidelines in oncology on prostate cancer. J Natl Compr Canc Netw 2010;8:145. [Crossref] [PubMed]
- Heidenreich A, Bellmunt J, Bolla M, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localised disease. Eur Urol 2011;59:61-71. [Crossref] [PubMed]
- Nath A, Singh J, Vendan SE, et al. Elevated level of prostate specific antigen among prostate cancer patients and high prevalence in the Gangetic zone of Bihar, India. Asian Pac J Cancer Prev 2012;13:221-3. [Crossref] [PubMed]
- Greene KL, Albertsen PC, Babaian RJ, et al. Prostate specific antigen best practice statement: 2009 update. J Urol 2009;182:2232-41. [Crossref] [PubMed]
- Crawford ED, Rove KO, Trabulsi EJ, et al. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. J Urol 2012;188:1726-31. [Crossref] [PubMed]
- Elshafei A, Chevli KK, Moussa AS, et al. PCA3-based nomogram for predicting prostate cancer and high grade cancer on initial transrectal guided biopsy. Prostate 2015;75:1951-7. [Crossref] [PubMed]
- de Kok JB, Verhaegh GW, Roelofs RW, et al. DD3PCA3, a very sensitive and specific marker to detect prostate tumors. Cancer Res 2002;62:2695-8. [PubMed]
- Hessels D, Gunnewiek JMK, van Oort I, et al. DD3PCA3-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol 2003;44:8-15. [Crossref] [PubMed]
- Wei JT, Feng Z, Partin AW, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol 2014;32:4066. [Crossref] [PubMed]
- Ruffion A, Devonec M, Champetier D, et al. PCA3 and PCA3-based nomograms improve diagnostic accuracy in patients undergoing first prostate biopsy. Int J Mol Sci 2013;14:17767-80. [Crossref] [PubMed]
- Vlaeminck-Guillem V, Devonec M, Champetier D, et al. Urinary PCA3 to predict prostate cancer in a cohort of 1015 patients. Prog Urol 2015;25:1160-8, e1-8.
- Zheng K, Dou Y, He L, et al. Improved sensitivity and specificity for prostate cancer diagnosis based on the urine PCA3/PSA ratio acquired by sequence-specific RNA capture. Oncol Rep 2015;34:2439-44. [Crossref] [PubMed]
- Altintas DM, Allioli N, Decaussin M, et al. Differentially expressed androgen-regulated genes in androgen-sensitive tissues reveal potential biomarkers of early prostate cancer. PloS one 2013;8:e66278. [Crossref] [PubMed]
- Zhou Y, Li Y, Li X, et al. Urinary biomarker panel to improve accuracy in predicting prostate biopsy result in Chinese men with PSA 4–10 ng/mL. BioMed Res Int 2017;2017.
- Wang FB, Chen R, Ren SC, et al. Prostate cancer antigen 3 moderately improves diagnostic accuracy in Chinese patients undergoing first prostate biopsy. Asian J Androl 2017;19:238. [Crossref] [PubMed]
- Reebye V, Frilling A, Habib NA, et al. Intracellular adaptor molecules and AR signalling in the tumour microenvironment. Cell Signal 2011;23:1017-21. [Crossref] [PubMed]


