Development and validation of an integrative methylation signature and nomogram for predicting survival in clear cell renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) is a common renal malignancy that originates from renal tubular epithelial cells, accounting for 2–3% of all adult malignant tumors and 85−90% of primary malignant renal tumors (1). Clear cell RCC (ccRCC) is the most common subtype of RCC and accounts for 70% of this disease, with increasing incidence and mortality rates worldwide (2). The identification of accurate predictors of clinical outcomes is vital for the treatment management of ccRCC. Currently, the assessment for the prognosis of ccRCC is mainly based on the tumor, nodes, metastasis (TNM) staging system and Fuhrman grade (3). However, clinical outcomes among ccRCC patients with the same clinical stage may differ significantly, meaning that these systems still cannot accurately predict prognosis in ccRCC patients. Therefore, novel molecular biomarkers are urgently needed for the early detection and precise survival prediction for ccRCC.
During the past few decades, genetic or epigenetic alterations have been recognized as playing a crucial role in the occurrence and development of various types of cancers (4,5). DNA methylation is an epigenetic modification with high potential in revealing cancer initiation, monitoring therapy response, and predicting the clinical outcome (6). Growing evidence suggests DNA methylation is highly involved in the initiation and progression of ccRCC and could thus serve as a useful biomarker for predicting the prognosis (7). Moreover, accumulating new evidence has revealed that a biomarker signature consisting of several methylated gene members may be more qualified due to its higher prediction power than single molecules, as it integrates the effect of multiple genes and accordingly provides a more comprehensive prediction of clinical behavior (8). Previous epigenetic studies have attempted to explore a series of frequently methylated genes in ccRCC; however, the classification may not be detailed enough and has not been sufficiently proven to allow clinicians to reach more informed decision-making for treatment management and survival outcome improvement (9).
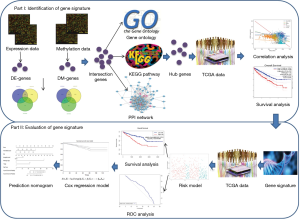
This study therefore developed and established an integrated and comprehensive methylation signature with well-defined risk scores for ccRCC. This might serve as a foundation for understanding the mechanism of ccRCC involved in methylation and provide novel biomarkers for the prognosis or treatment of ccRCC. The schematic pipelines of this study are presented in Figure 1. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tau-19-853).
Methods
Data source
Two ccRCC gene expression profiles (GSE15641 and GSE53757) were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repositories (http://www.ncbi.nlm.nih.gov/geo/) (10,11). A total of 32 ccRCC tumor specimens and 23 adjacent normal tissues were available for GSE15641, while 72 pairs of ccRCC and matched normal tissues were included for GSE53757. The DNA methylation data of ccRCC (GSE70303) were also retrieved from the GEO database, which contained 12 pairs of ccRCC and matched adjacent normal tissues (12). Level 3 methylation and expression data, along with the corresponding clinical information for ccRCC patients, were obtained from The Cancer Genome Atlas (TCGA) database (13).
Data processing
For the gene expression data, the ‘affy’ package in the R language environment was applied to normalize and correct the expression values (14). When multiple probes annotated a gene symbol, the average expression value of these probes was then calculated to represent the expression level of this gene. For the methylation data, we used the probes located in the gene promoter region as the methylation level of the gene. The methylation profile data were normalized using quantile normalization in the R language environment, and then the gene promoter methylation status was evaluated by calculating the mean values of a given promoter region. Accordingly, the differentially expressed genes and methylated genes were identified based on the Limma package in R Statistical Program (15). P value <0.05 and |fold-change| >2 were considered as statistically significant. Then, a list of genes was identified by the intersection of the differentially expressed genes with the differentially methylated genes and used for further analysis.
Functional enrichment analysis
Functional enrichment analysis was performed with these identified genes. In this study, the Search Tool for the Retrieval of Interacting Genes (STRING) database was applied. STRING is a robust database containing comprehensive protein-protein association data from functional discovery in genome-wide experimental datasets. STRING implements well-known classification systems, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (16-18). Thus, the significantly enriched GO terms and KEGG pathways were identified based on STRING. The GO analysis was divided into 3 different levels including molecular function, cell component, and biological processes. P value <0.05 and gene count ≥2 were selected as the cut-off criteria.
Protein-protein interaction (PPI) network analysis
To further understand the associations among the screened gene list and to identify more important genes, we performed a PPI network analysis. We first uploaded the gene list to the STRING database and then set up the PPI network with the identified genes by integrating the data retrieved from the STRING database. The PPI network was then visualized by using the Cytoscape tool. The hub genes were named based on combined scores (>0.7) and connection numbers (>8).
Correlation and prognosis analysis
Based on the Spearman’s correlation coefficient, the correlations between promoter methylation levels and the corresponding gene expression levels were investigated with methylation data and the matched gene expression data. The genes calculated with the negative correlation coefficient values were chosen for further analysis. After that, the Kaplan-Meier method with log-rank test was used to evaluate the correlation between the selected genes and the overall survival (OS) of ccRCC patients (19,20).
Prognostic model construction and evaluation
A prognostic gene methylation signature was constructed based on the genes identified from the above step. Then, a receiver operating characteristic (ROC) curve was constructed with the ratio value for each marker in the prognostic signature, and the area under the curve (AUC) was calculated for evaluating the diagnostic power of each marker in detecting ccRCC patients (21). A survival risk score model was set up to better assess the capability of the gene signature for predicting OS. With a median risk score as the cutoff, the patients were divided into 2 groups, including the high-risk (risk score > cutoff) groups and low-risk groups (risk score < cutoff). Then the AUC of the ROC curve was applied to assess the performance of the risk groups.
Univariate and multivariate Cox regression analysis
To identify whether the gene methylation signature and other clinical variables were independent factors in the survival outcome of ccRCC patients, univariate Cox proportional hazard analysis and multivariate Cox regression analysis were conducted by using the survival package in R language environment (22). Only the significant variables (P value <0.05) in the univariate model were selected to conduct the multivariate logistic regression analysis.
Construction of a predictive nomogram
Finally, a prediction nomogram was constructed based on the significant prognostic factors of the multivariate Cox regression analysis by RMS package in the R Statistical Program (23). The calibration curve was generated to evaluate the calibration ability of the nomogram. The concordance index (C-index) was applied to evaluate the discriminative capacity of the nomogram (24). The C-index ranges from 0.5 to 1.0 and a higher C-index were considered to have superior discriminative capacity for prognosis. A C-index value ≥0.70 was suggested to possess an accurate prognostic prediction. Every variable in the nomogram was assigned a score. A risk classification system was developed based on the total scores of each ccRCC patient by using the nomogram to stratify ccRCC patients into 3 prognostic groups: the low-, intermediate-, and high-risk groups. Then, the Kaplan-Meier method with log-rank test was used to analyze the difference in OS among the 3 different risk groups.
Results
Screening of differentially expressed and methylated genes
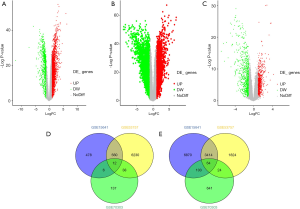
Based on the selected sample data of GSE15641, a total of 11,507 differentially expressed genes were found between ccRCC tissues and normal tissues, including 10,451 up-regulated and 1,056 down-regulated genes. For GSE53757, a total of 5,326 up-regulated and 6,840 down-regulated genes were found. We obtained 3,478 up-regulated and 639 down-regulated genes by combining the 2 datasets. According to the methylation dataset, a total of 1,025 differentially methylated genes were screened, among which 193 were hypermethylated, and 832 were hypomethylated. The volcano plots for differentially expressed and methylated genes are plotted in Figure 2. After crosstalk, a total of 76 genes, including 12 hypermethylated down-regulated and 64 hypomethylated up-regulated genes, were identified and selected for further analysis. The Venn diagrams for differentially expressed and methylated genes are also presented in Figure 2.
Integrative functional analysis results
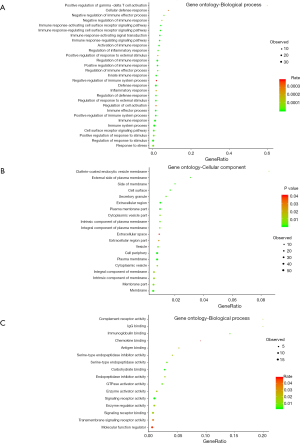
To further understand the biological functions of these methylation related genes, we performed an integrative functional enrichment analysis of those identified 76 genes. The results of the GO analysis are shown in Figure 3. At the molecular function level, the identified genes significantly correlated with some key molecule activity, including complement receptor, signaling receptor, and GTPase activator and binding function such as IgG binding, immunoglobulin binding, and chemokine binding. At the cell component level, these genes were highly associated with some critical cell structures. For the biological processes, the identified genes were linked with the regulation of various biological activities. The results of KEGG analysis showed that the identified genes were highly involved in several signaling pathways, including staphylococcus aureus infection, osteoclast differentiation, and natural killer cell-mediated cytotoxicity.
PPI network construction and identification of hub genes
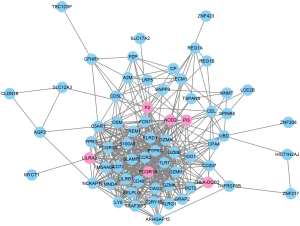
To further evaluate the associations among the identified 76 genes, we established a PPI network by integrating the data retrieved from the STRING database. The PPI network from the STRING for the 76 genes was visualized by using the Cytoscape tool (Figure 4). Based on the combined scores (>0.7) and connection numbers (>8), 6 genes were identified as hub genes, including F2, FCGR1A, HLA-DQB2, LILRA2, NOD2, and PI3.
Correlation analysis and identification of signature genes
We first explored the correlation between methylation levels and gene expression. Spearman’s rank coefficient was calculated for determining the correlation between the methylation and hub gene expression data. The results indicated that FCGR1A, F2, LILRA2, and NOD2 correlated negatively with the level of methylation (P<0.05) while HLA-DQB2 and PI3 had no significant associations with methylation level (Figure 5). The 4 genes negatively associated with methylation levels were selected for further analysis.
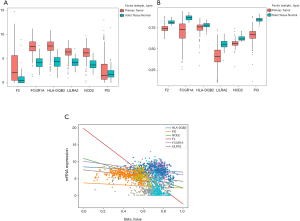
To identify the selected 4 genes which would be potentially related to the OS of ccRCC patients, the Kaplan-Meier analysis and log-rank test were then performed to assess the association between gene expression and patients’ survival. The results showed that FCGR1A, F2, and NOD2 were negatively correlated with the OS of ccRCC patients, while LILRA2 revealed no significant relationship with the OS of ccRCC patients. The survival curves for the 4 genes are plotted in Figure 6. The 3 genes were then selected as candidates for further analysis.
The diagnostic power of the signature genes in detecting ccRCC patients
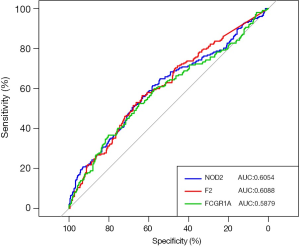
The capabilities of FCGR1A, F2, and NOD2 in discriminating ccRCC patients from normal cases were further evaluated using the ROC analysis. The ROC curve (Figure 7) was plotted, and the AUCs of the 3 genes were 0.6054 (NOD2), 0.6088 (F2), and 0.5879 (FCGR1A), revealing that these 3 genes had good distinguishing ability and may serve as the potential biomarkers for the early detection of ccRCC.
Construction and evaluation of the predictive gene signature
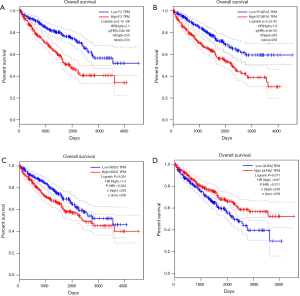
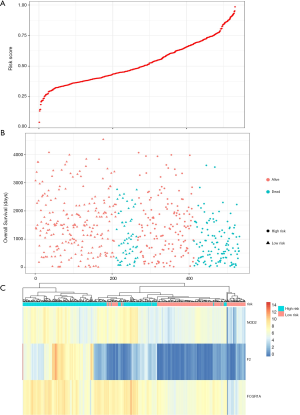
Prognostic methylation of differentially expressed genes (MDEGs) signature based on FCGR1A, F2, and NOD2 was constructed by integrating the data of the 3 candidates and corresponding estimated regression coefficient. Next, the risk score of every ccRCC patient was calculated and ranked. The patients were stratified based on the median risk score into high-risk (n=267) and low-risk (n=266) groups under each risk scoring system. The risk score distribution, survival score distribution, and the expression heat-map were plotted in Figure 8. Then, survival analysis was conducted with the Kaplan-Meier method and log-rank statistical test. The OS rates of ccRCC patients were 60% in the high-risk group and 75.8% in the low-risk group (Figure 9). The patients with ccRCC in the high-risk group had a significantly shorter OS than those in the low-risk group (HR, 2.46; 95% CI, 1.63–3.71; P<0.001). Therefore, the result revealed that patients in the high-risk group were correlated with a significantly worse prognosis than patients in the low-risk group.
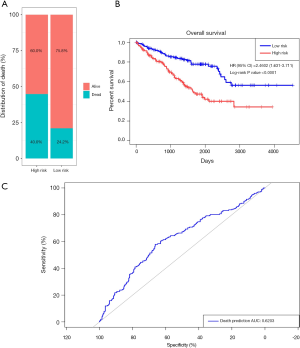
The sensitivity and specificity of the signature for predicting OS were assessed by using ROC analysis to evaluate the predictive power of the methylation genes signature. According to the ROC analysis results (Figure 9), the three-gene methylation signature achieved the overall AUC of 0.6203, showing its moderate diagnosis power. Thus, it may be used as a novel and moderately correct prognostic biomarker for predicting the survival outcome of ccRCC patients.
Univariate and multivariate Cox regression of the predictive signature
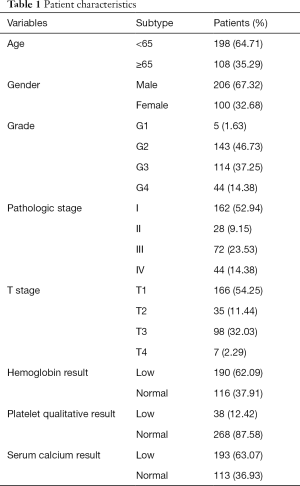
A total of 306 patients clinically and pathologically diagnosed with ccRCC in TCGA were used to construct the predictive model for the prognosis of ccRCC patients. The detailed characteristics are presented in Table 1. Considering the clinical factors including age, sex, pathologic stage, T stage, tumor grade, hemoglobin result, platelet qualitative result, and serum calcium result, univariate and multivariate Cox regression analyses were applied for evaluating the effect of the three-gene methylation signature (high risk vs. low risk) on OS. The univariate analysis indicated that age (HR =1.8, P=0.003), tumor grade (HR =2.3, P<0.001), pathologic stage (HR =3.47, P<0.001), T stage (HR =2.76, P<0.001), hemoglobin result (HR =0.54, P=0.006), platelet qualitative result (HR =0.61, P=0.059), and three-gene methylation signature (HR =2.46, P<0.001) were correlated with OS in ccRCC patients. When integrating the independent factors into multivariate Cox regression analysis, age (HR =1.58, P=0.023), pathologic stage (HR =5.77, P<0.001), T stage (HR =0.44, P=0.04), hemoglobin result (HR =0.58, P=0.017), and three-gene methylation signature (HR =1.97, P=0.003) remained as independent prognostic factors for OS in ccRCC patients (Table 2).
Full table

Full table
Building and assessment of a predictive nomogram
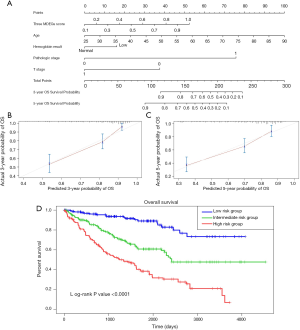
To build a model that could serve as an individual’s prognostic predictor, a nomogram was set up for predicting the 3- and 5-year OS by incorporating 5 independent covariates including age, pathologic stage, T stage, hemoglobin result, and three-gene methylation signature. The predictive nomogram is plotted in Figure 10. C-index was applied to assess the discrimination power of the model. As a result, the C-index for predicting OS in the nomogram was 0.762, which shows good predictive ability. The calibration curves of the nomogram revealed excellent consistency between the values of prediction and observation in 3-, and 5-year OS probability (Figure 10). The total score was calculated by adding the individual scores of all the selected variables. Then, we divided the ccRCC patients into 3 different risk groups, including the high-, median-, and low-risk groups based on the scores of the nomogram with the cutoff value of 172 and 130. Kaplan-Meier curves for OS of ccRCC patients from TCGA were developed for each risk group, showing there was a significant difference among the 3 risk groups (Figure 10). We further compared the predictive OS power between the established nomogram and the prognostic ability of the independent variables. Promisingly, the C-index of the nomogram was significantly higher than those of the independent variables for OS prediction (C-index: 0.762 vs. 0.746; P<0.05), revealing the constructed nomogram had the better discriminative capacity for predicting OS of ccRCC patients.
Discussion
Despite the advancements of treatment management and cancer surveillance of ccRCC, the prognosis of this disease is still poor. Current prognostic methods for ccRCC are still not sufficient for accurate prediction and individualized treatment. Genetic or epigenetic biomarkers have opened a window for the diagnosis, therapy, and prognosis of ccRCC as they can better reveal the underlying information of cancer than traditional markers. It is well established that alterations in DNA methylation play a vital part in the occurrence and progression of ccRCC and provide clinically viable biomarkers for early diagnosis and precise treatment of ccRCC. In this study, we systematically and comprehensively screened and showed a methylation signature associated with the prognosis of ccRCC through an integrated biomarker discovery phase. In addition, we developed a predictive model based on the identified methylation signature that may be useful for improving the clinical management of ccRCC.
We firstly found differentially expressed and methylated genes from 3 datasets. After the intersection, 76 genes were obtained for constructing the PPI network. Then 6 genes were found as hub genes, including F2, FCGR1A, HLA-DQB2, LILRA2, NOD2, and PI3. The correlation and survival analysis revealed that FCGR1A, F2, and NOD2 negatively correlated with the level of methylation and the OS of ccRCC patients. Following that, a prognostic gene methylation signature was constructed based on FCGR1A, F2, and NOD2. FCGR1A has been identified as an interferon-inducible gene that is highly expressed by myeloid cells, such as macrophages and neutrophils (25). Notably, FCGR1A has also been demonstrated to possess a diagnostic and prognostic potential in a series of diseases, including antibody-mediated rejection, tuberculosis, and triple-negative breast cancer (26-28). It is widely acknowledged that F2 is a coagulation factor that is proteolytically cleaved to generate thrombin in the original process of the coagulation cascade leading to the stemming of blood loss. F2 also plays fundamental and pleiotropic roles in maintaining vascular integrity during development and postnatal life (29,30). NOD2, an intracellular pattern recognition receptor, plays its role by sensing bacterial peptidoglycan-conserved motifs in the cytosol and stimulating host immune response, including in epithelial and immune cells (31). Recently gathered evidence has indicated that NOD2 is highly involved in host defense against infection and the control of inflammation (32). One previous study discovered that NOD2 was more highly expressed in human ccRCC tissue than in adjacent healthy tissue, and modulation of NOD2 receptors might provide a molecular therapeutic approach in ccRCC (33). However, the underlying mechanism of the occurrence and development of FCGR1A, F2, and NOD2 is still poorly understood, and it is thus worth further exploring the mechanisms of these molecules that contribute to ccRCC carcinogenesis. Moreover, further characterization of these molecules may offer new insights into the individualized management of ccRCC and might contribute to the identification of potential therapeutic targets for ccRCC.
A prognosis-related risk scoring system was established with the signature genes to evaluate the predictive power of the prognostic signature. Based on the risk score, ccRCC patients were divided into low- and high-risk groups with significantly different OS. Moreover, ROC curves demonstrated the high specificity and sensitivity of the prognostic signature in the survival prediction of patients with ccRCC. The results showed that the prognostic signature might have the potential for scheduling treatment strategies and guiding individualized follow-up for ccRCC patients. For example, we suggest that patients identified as high-risk should accept more intensive follow-up or therapy.
It is generally accepted that ccRCC is a heterogeneous disease, and a variety of confounding factors contribute to the establishment and development of this kind of cancer. A single marker is unlikely to be very valuable for survival prediction. Therefore, several clinical factors, along with the signature genes, were enrolled in the univariate analysis and were demonstrated to be significantly associated with OS in ccRCC patients. Moreover, when integrated into the multivariate Cox regression analysis, the signature genes remained independent prognostic factors for OS in ccRCC patients. These clinical factors and the predictive signature could be combined to serve as predictive substrates in predicting the survival of ccRCC patients.
In recent years, a nomogram has become a promising component in assessing survival or specific outcomes by showing visual graphical interfaces. Moreover, the nomogram has superior predictive properties than conventional American Joint Committee on Cancer (AJCC) TNM staging under complicated clinical conditions and thus could play more critical roles in modern medical decision-making (34,35). In our study, we set up a nomogram consisting of 5 independent covariates generated from a multivariate Cox regression analysis for OS prediction of ccRCC patients. The predictive performance of the nomogram was shown to be effective in TCGA cohorts. Thus, our nomogram might facilitate decision-making as a simple, visual tool for predicting the prognosis of ccRCC patients.
Although a series of biomarkers has been identified for ccRCC, prognostic markers of ccRCC that could provide valuable information regarding prognosis and treatment options at diagnosis are still a long way off. A study reported that herpes virus entry mediator (HVEM) might serve as a promising and independent adverse predictor of survival outcomes in ccRCC patients. Another study identified tumor suppressor candidate 3 (TUSC3) to be a promising tumor biomarker for the early diagnosis and prognosis of ccRCC (36). High expressions of BMP1 were significantly associated with poor prognosis and may serve as a potential prognostic factor and therapeutic target (37). Moreover, a predictive signature was constructed based on the expression of 5 genes and was demonstrated to be an independent prognostic factor for ccRCC (38). These studies reflect the potential of the markers identified in clinical application. Most of these studies were, however, restricted with an isolated and static mode that focused on limited molecules. Our previous studies have proven that combination markers outperform single molecules in cancer characterization (39,40). Thus, combining methylation markers with other biomarkers may provide a new alternative for clinical application. A more complicated model that integrates the more useful parameters will make the prognostic model more powerful.
It is important to acknowledge our study has several limitations. Firstly, the sample sizes of the datasets that were used for identifying the differentially expressed and methylated genes were still limited. Secondly, due to the lack of information, some prognostic variables were not included in the nomogram, but they may be useful in modifying the nomogram to provide more accurate prognoses in clinical practice. Thirdly, the mechanisms behind the prognostic values of these methylation genes in ccRCC are still poorly understood, as we have not performed biological experiments that may provide vital information to further improve our understanding of their functional roles. Finally, although the predictive model was verified to be useful by published data, it has not yet been checked prospectively in a clinical trial. Regardless of these limitations, the significant and consistent correlations of our prognostic signature genes and nomogram with OS indicates that they may act as convenient and accurate prognosis prediction tools for ccRCC, and are worthy of further investigation.
Conclusions
To conclude, we developed and validated a three-gene methylation signature for predicting the prognosis of ccRCC. A nomogram, including the signature, could aid the prediction of individual OS and may help clinicians in decision-making for individualized treatment. Future clinical studies and biological experiments should be performed to further evaluate the predictive power of the three-gene methylation signature and examine their functional mechanisms in the pathogenesis and development of ccRCC.
Acknowledgments
We thank the AME Editing Service (http://editing.amegroups.cn/#editing) for improving our paper.
Funding: This work was supported by the National Natural Science Foundation of China (grant no. 81773221) for JZ, the Natural Science Foundation of Jiangsu Province (grant no. BK20161222) for JZ. It was also funded by Suzhou Science and Technology Planed Projects (grant no. SYS201629, SS201857) for JZ and the grant for Key Young Talents of Medicine in Jiangsu (grant No. QNRC2016875) for JZ.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tau-19-853
Data Sharing Statement: Available at http://dx.doi.org/10.21037/ tau-19-853
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-19-853). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers 2017;3:17009. [Crossref] [PubMed]
- Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ 2014;349:g4797. [Crossref] [PubMed]
- Barata PC, Rini BI. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J Clin 2017;67:507-24. [Crossref] [PubMed]
- Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med 2019;25:403-18. [Crossref] [PubMed]
- Thavaneswaran S, Rath E, Tucker K, et al. Therapeutic implications of germline genetic findings in cancer. Nat Rev Clin Oncol 2019;16:386-96. [Crossref] [PubMed]
- Michalak EM, Burr ML, Bannister AJ, et al. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat Rev Mol Cell Biol 2019;20:573-89. [Crossref] [PubMed]
- Capitanio U, Bensalah K, Bex A, et al. Epidemiology of Renal Cell Carcinoma. Eur Urol 2019;75:74-84. [Crossref] [PubMed]
- Van Hoeck A, Tjoonk NH, van Boxtel R, et al. Portrait of a cancer: mutational signature analyses for cancer diagnostics. BMC Cancer 2019;19:457. [Crossref] [PubMed]
- Wang J, Zhang Q, Zhu Q, et al. Identification of methylation-driven genes related to prognosis in clear-cell renal cell carcinoma. J Cell Physiol 2020;235:1296-308. [Crossref] [PubMed]
- Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res 2005;11:5730-9. [Crossref] [PubMed]
- von Roemeling CA, Radisky DC, Marlow LA, et al. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res 2014;74:4796-810. [Crossref] [PubMed]
- Becket E, Chopra S, Duymich CE, et al. Identification of DNA Methylation-Independent Epigenetic Events Underlying Clear Cell Renal Cell Carcinoma. Cancer Res 2016;76:1954-64. [Crossref] [PubMed]
- International Cancer Genome Consortium, Hudson TJ, Anderson W, et al. International network of cancer genome projects. Nature 2010;464:993-8. [Crossref] [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. Affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47:D330-8. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Schultz LR, Peterson EL, Breslau N. Graphing survival curve estimates for time-dependent covariates. Int J Methods Psychiatr Res 2002;11:68-74. [Crossref] [PubMed]
- Koletsi D, Pandis N. Survival analysis, part 2: Kaplan-Meier method and the log-rank test. Am J Orthod Dentofacial Orthop 2017;152:569-71. [Crossref] [PubMed]
- Obuchowski NA, Bullen JA. Receiver operating characteristic (ROC) curves: review of methods with applications in diagnostic medicine. Phys Med Biol 2018;63:07TR1.
- Lusa L, Miceli R, Mariani L. Estimation of predictive accuracy in survival analysis using R and S-PLUS. Comput Methods Programs Biomed 2007;87:132-7. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Wolbers M, Koller MT, Witteman JC, et al. Prognostic models with competing risks: methods and application to coronary risk prediction. Epidemiology 2009;20:555-61. [Crossref] [PubMed]
- Jenum S, Bakken R, Dhanasekaran S, et al. BLR1 and FCGR1A transcripts in peripheral blood associate with the extent of intrathoracic tuberculosis in children and predict treatment outcome. Sci Rep 2016;6:38841. [Crossref] [PubMed]
- Van Loon E, Gazut S, Yazdani S, et al. Development and validation of a peripheral blood mRNA assay for the assessment of antibody-mediated kidney allograft rejection: A multicentre, prospective study. EBioMedicine 2019;46:463-72. [Crossref] [PubMed]
- Gebremicael G, Kassa D, Quinten E, et al. Host Gene Expression Kinetics During Treatment of Tuberculosis in HIV-Coinfected Individuals Is Independent of Highly Active Antiretroviral Therapy. J Infect Dis 2018;218:1833-46. [Crossref] [PubMed]
- Jiang YZ, Liu YR, Xu XE, et al. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Cancer Res 2016;76:2105-14. [Crossref] [PubMed]
- Gigante B, Bennet AM, Leander K, et al. The interaction between coagulation factor 2 receptor and interleukin 6 haplotypes increases the risk of myocardial infarction in men. PLoS One 2010;5:e11300. [Crossref] [PubMed]
- Gigante B, Vikstrom M, Meuzelaar LS, et al. Variants in the coagulation factor 2 receptor (F2R) gene influence the risk of myocardial infarction in men through an interaction with interleukin 6 serum levels. Thromb Haemost 2009;101:943-53. [Crossref] [PubMed]
- Keestra-Gounder AM, Tsolis RM. NOD1 and NOD2: Beyond Peptidoglycan Sensing. Trends Immunol 2017;38:758-67. [Crossref] [PubMed]
- Mukherjee T, Hovingh ES, Foerster EG, et al. NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophys 2019;670:69-81. [Crossref] [PubMed]
- Mey L, Jung M, Roos F, et al. NOD1 and NOD2 of the innate immune system is differently expressed in human clear cell renal cell carcinoma, corresponding healthy renal tissue, its vasculature and primary isolated renal tubular epithelial cells. J Cancer Res Clin Oncol 2019;145:1405-16. [Crossref] [PubMed]
- Dong D, Tang L, Li ZY, et al. Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer. Ann Oncol 2019;30:431-8. [Crossref] [PubMed]
- Wang S, Yang L, Ci B, et al. Development and Validation of a Nomogram Prognostic Model for SCLC Patients. J Thorac Oncol 2018;13:1338-48. [Crossref] [PubMed]
- Yan Y, Chen Z, Liao Y, et al. TUSC3 as a potential biomarker for prognosis in clear cell renal cell carcinoma. Oncol Lett 2019;17:5073-9. [PubMed]
- Xiao W, Wang X, Wang T, et al. Overexpression of BMP1 reflects poor prognosis in clear cell renal cell carcinoma. Cancer Gene Ther 2020;27:330-40. [Crossref] [PubMed]
- Pan Q, Wang L, Zhang H, et al. Identification of a 5-Gene Signature Predicting Progression and Prognosis of Clear Cell Renal Cell Carcinoma. Med Sci Monit 2019;25:4401-13. [Crossref] [PubMed]
- Peng Q, Feng Z, Shen Y, et al. Integrated analyses of microRNA-29 family and the related combination biomarkers demonstrate their widespread influence on risk, recurrence, metastasis and survival outcome in colorectal cancer. Cancer Cell Int 2019;19:181. [Crossref] [PubMed]
- Peng Q, Shen Y, Lin K, et al. Identification of microRNA-92a and the related combination biomarkers as promising substrates in predicting risk, recurrence and poor survival of colorectal cancer. J Cancer 2019;10:3154-71. [Crossref] [PubMed]