Nomograms for predicting overall and cancer-specific survival in patients with papillary renal cell carcinoma: a population-based study using SEER database
Introduction
Renal cell carcinoma (RCC), as the most common type of kidney cancers, is mainly classified into clear cell RCC (ccRCC) and non-ccRCC according to the histology (1). ccRCC was the most common subtype of RCC, accounting for 75–80% of the total diagnosed cases, while papillary RCC (pRCC) ranked the second, accounting for approximately 10–15% of the total diseases (2,3). Further, pRCC could be divided into two major subtypes according to histopathological features. Type I tumors are usually with thin basophilic papillary cells, while type II pRCCs are composed of thicker nipples and eosinophilic cytoplasm (4).
Currently, the prognosis of pRCC remained poor, and there were still no effective methods for the treatment of advanced pRCC (5). Therefore, it was essential to identify related prognostic factors for improving the survival of pRCC at early stage. Traditionally, TNM stage was regarded as one of the most important prognostic factors in various cancers (6). However, it was not sufficient to cover the biological characteristics of cancer and predict survival outcomes (7). In addition, other clinical variables such as age, gender, ethnicity, grade, surgical treatment, adjuvant therapy, and molecular characteristics may generate influence on the prognosis of cancer patients (8). Nevertheless, the prognostic value of these parameters in pRCC remained inconsistent and even doubtful.
In recent years, there have been a lot of tools used to predict the survival outcomes in numerous cancers (9-11). Of the available tools, nomogram is currently one of the most effective and accurate methods for predicting the prognosis of cancer patients (12). The aim of this retrospective study was to explore the clinicopathological features associated with the prognosis of pRCC and to construct nomograms to predict survival on account of these features.
Methods
Patients
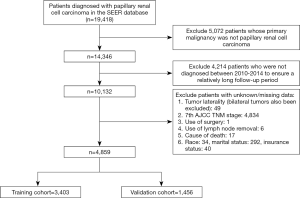
Primary data of patients with pRCC were obtained from the Surveillance, Epidemiology, and End Results (SEER) database (http://seer.cancer.gov/) utilizing the SEER*Stat software [Version 8.3.6; National Cancer Institute, Bethesda, United States (US)]. It is a public and population-based database which covers approximately 30% of the US population. The inclusion criteria of this study were as follows: (I) diagnosed as pRCC (International Classification of Diseases for Oncology: 8260/3) with positive histology, (II) year at diagnosis was from 2010 to 2014 to ensure a relatively long follow-up period, (III) complete data were available with active follow-up. Additionally, the exclusion criteria were as follows: (I) missing/unknown data in following variables: age, sex, race, American Joint Committee on Cancer (AJCC) 7th edition TNM stage, tumor laterality (bilateral tumors also been excluded), surgery, radiation, chemotherapy, follow-up time, insurance status, marital status, survival outcomes and so on, (II) pRCC was not the first primary malignancy, (III) type of reporting source was autopsy only or death certificate only.
Primary data were reviewed respectively by two independent investigators (Haicui Yan and Xiyi Wei) to extract the clinical characteristics and survival outcomes of the enrolled patients. Variables included age, sex, race, tumor laterality, AJCC 7th edition TNM stage, the use of radiation, chemotherapy, surgery, lymph node removal (LNR), survival months, survival status, insurance status and marital status. The primary endpoints were overall survival (OS) and cancer-specific survival (CSS). Survival time was calculated from the date of diagnosis to the date of death from pRCC (defined as CSS) or any disease cause (defined as OS). Lastly, use of SEER was exempt from Institutional Review Board (IRB).
Training and validation group
In order to develop the prognostic nomograms and undergo further external validation, all of the enrolled patients were divided into training group and validation group randomly at a ratio of 7:3 by using random-number generation method. Finally, chi-square test was utilized to make comparisons in basic characteristics between two groups.
Statistical analysis
Uni- and multivariate Cox regression analyses were conducted to explore the prognostic factors which affect OS and CSS significantly. Additionally, hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) of selected factors were calculated. According to the results of multivariate Cox regression analysis, predictive nomograms for 3- and 5-year CSS and OS were developed. In the training group, survival curves for different variables were produced by Kaplan-Meier (KM) analyses and were compared utilizing the log-rank test.
To assess the predictive ability and accuracy of the nomograms, discrimination and calibration of the nomograms were measured in two groups. The area under receiver operating characteristic (ROC) curve (defined as AUC) (13,14) and Harrell’s concordance index (C-index) (15) were applied to assess the discrimination. The AUC and C-index range from 0.5 to 1.0, with 0.5 suggesting the total chance and 1.0 suggesting a perfect discrimination ability (16). Calibration curves were performed to identify the consistency between the observed survival and the predicted survival.
The Cox analysis and chi-square test were conducted via SPSS 23.0 software (SPSS Inc, Chicago, IL, US). Survival and RMS package were used to develop and validate the predictive nomograms via RStudio software (Version 1.2.5001). Two-sided P<0.05 was considered to be statistically significant during the whole analysis process.
Results
Basic characteristics
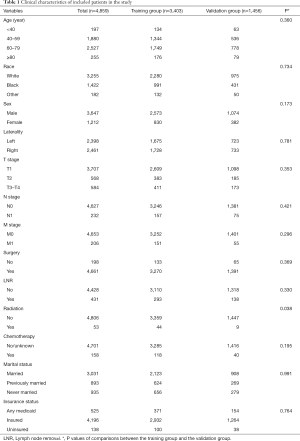
A total of 4,859 patients with pRCC diagnosed between 2010 and 2014 were included in this study (selection flow chart was in Figure 1. After randomly grouping, 3,403 patients were included in the training cohort and the remaining 1,456 patients were in the validation cohort. The training group was used for the development and internal validation of the predictive nomograms while the validation group was assigned for the external validation.
In the total cohort, most of the patients were male (75.06%), white (66.99%), and had a tumor in early T stage (76.29%) and without metastasis (95.76%). In terms of tumor laterality, no significant difference was detected between these two groups. Most patients had undergone surgery (95.93%) and a small number of patients had received chemotherapy (3.25%) and radiotherapy (1.29%). Detailed clinical information and comparisons between two groups was summarized in Table 1. There were no significant differences in race, age, sex, tumor laterality, TNM stage, insurance status, marital status, or use of surgery/chemotherapy/radiotherapy/LNR (all P>0.05). However, patients with radiotherapy in the training group were more than those in the validation group significantly (P=0.038).
Full table
Cox analyses, KM analyses and nomograms construction
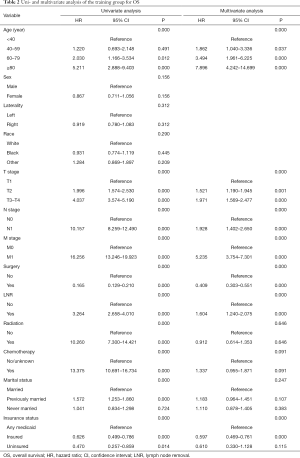
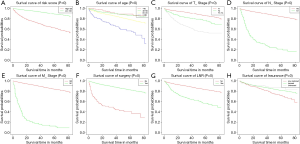
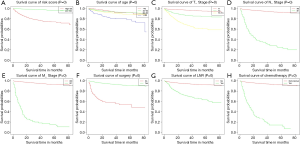
As shown in Table 2 and Table 3, in univariate Cox analysis, 13 variables were enrolled including age, race, sex, tumor laterality, T stage, N stage, M stage, use of surgery/chemotherapy/radiotherapy/LNR, insurance status and marital status. Eventually, seven factors (age, T stage, N stage, M stage, the use of surgery, LNR and insurance status) were significantly related to OS and seven factors (age, T stage, N stage, M stage, the use of surgery, chemotherapy and LNR) were significantly associated with CSS. Finally, KM survival curves for OS and CSS were generated to learn the actual effect of different variables (Figures 2,3).
Full table
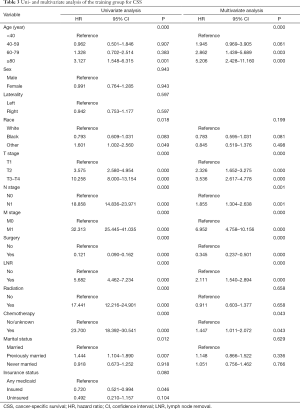
Full table
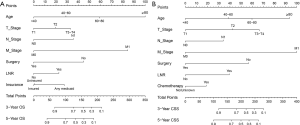
Based on the results of multivariate Cox analysis, nomograms to predict 3- and 5-year OS and CSS were conducted (Figure 4). A total point can be obtained by adding the score of each variable and the total point had its corresponding OS/CSS probabilities by the nomograms.
Nomogram validation
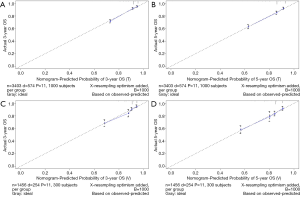
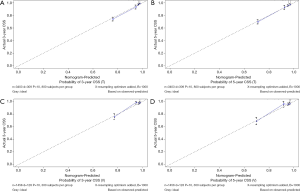
The C-index of the OS predictive model was 0.764 in the training group and 0.723 in the validation group. As to the CSS nomogram, it was 0.859 in the training group and 0.824 in the validation group, respectively. For OS, the 3- and 5-year AUCs were 0.779 and 0.752 in the training group and 0.749 and 0.722 in the validation group (Figure 5). For CSS, the 3- and 5-year AUCs were 0.871 and 0.844 in the training group and 0.853 and 0.822 in the validation group (Figure 6). These results suggested the predictive nomograms were with good discrimination performance. Furthermore, calibration curves for 3- and 5-year indicated a good consistency between the observed survival and the predicted survival in both in OS (Figure 7) and CSS (Figure 8).


Discussion
As mentioned above, RCC accounts for approximately 90% of kidney cancer (17). As one of the prominent subtypes of RCC, pRCC accounts for approximately 6–18% of renal tumors (17,18) and is the dominant histological subtype in pediatric RCC patients. Unfortunately, approximately 20% of pRCCs were found to be incidental, and with specific symptoms and prognostic factors (19,20). Therefore, it was crucial to determine related prognostic factors to improve the prognosis of patients with pRCC. Because the nomogram can predict survival risk by combining and quantifying the relative importance of various prognostic factors, it has been widely applied for clinical oncology assessment.
Our study demonstrated that age, T stage, N stage, M stage, surgery, LNR and insurance status were prognostic factors for OS. Moreover, age, T stage, N stage, M stage, use of surgery, chemotherapy and LNR were significantly associated with CSS. Nomograms were then developed based on these prognostic factors to predict 3- and 5-year OS and CSS rate in patients with pRCC. Both in the training and validation cohorts, the nomograms showed good predictive ability.
Chai et al. (21) demonstrated that age, T stage, N stage, M stage, and the use of surgery/chemotherapy played an important role in the prognosis of cancer patients, which was consistent with our results. In our study, patients over 80 years old and patients with higher TNM stages appeared to have lower OS and CSS rates. Moreover, the use of surgery may effectively reduce the risk of recurrence and death. In addition, we found that chemotherapy was a negative prognostic factor in our model. Conversely, various multi-center clinical trials have shown that patients with non-ccRCC had a good response rate to chemotherapy and had considerable efficacy (22-24). The adjuvant chemotherapy seemed to be a protective prognostic factor in their study. This might be attributed to the fact that considerable proportion of patients receiving chemotherapy were diagnosed with metastatic or advanced papillary cancer.
Furthermore, with regard to LNR, it seemed that lymph node ratio had become the primary prognostic factor for tumor outcomes and showed significant advantages in breast cancer, ovarian cancer, and cervical cancer (25-27). LNR might significantly reduce the risk of survival in tumor patients. Interestingly, our results provided different opinions, where LNR was a risk factor in our model. Meanwhile, several other studies also suggested that the role of lymph node dissection in RCC remains controversial (28-30). The most recent systematic review (28) concluded that although LNR could provide independent prognostic information, the current literature failed to recognize the therapeutic benefit in non-metastatic or metastatic RCC. We attributed the adverse value of LNR in our model to the truth that the lymph node metastasis may have occurred in these patients before the dissection, and thus showing a poor prognosis. Nevertheless, LNR may still play a role in some high-risk non-metastatic patients and a further prospective study was completely substantial.
Overall, our research had the following advantages. First, our research was based on the population-based cancer database (SEER database), which collected cancer cases from 18 regions of the US, effectively avoiding selection bias in single-center and small-sample studies. Secondly, our study was the first attempt to establish prognostic nomograms of pRCC, and further tested its effectiveness by modest validation and good calibration. However, some limitations should not be ignored. First, difference was detected in the use of LNR between two groups, which probably due to the poor number of people with LNR. Second, the cases included in the study were from retrospective cohorts. Larger prospective randomized controlled trials were needed to verify the accuracy of the model.
In conclusion, prognostic factors for pRCC patients were identified and further survival nomograms were developed to predict the 3- and 5-year OS and CSS probabilities, providing an effective tool to assess individualized survival rate of pRCC patients.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (No.81702520), Medical Research Project of Jiangsu Provincial Health and Family Planning Commission (No. H2018052), Research Project of Jiangsu Cancer Hospital (No. ZN201602), and the Young Talents Program of Jiangsu Cancer Hospital (No. 2017YQL-04).
Footnote
Conflict of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-19-807). XL serves as an unpaid Section Editor of Translational Andrology and Urology from Oct 2019 to Dec 2021. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Use of SEER is exempt from Institutional Review Board.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet 2009;373:1119-32. [Crossref] [PubMed]
- Patard JJ, Leray E, Rioux-Leclercq N, et al. Prognostic value of histologic subtypes in renal cell carcinoma: a multicenter experience. J Clin Oncol 2005;23:2763-71. [Crossref] [PubMed]
- Beck SD, Patel MI, Snyder ME, et al. Effect of papillary and chromophobe cell type on disease-free survival after nephrectomy for renal cell carcinoma. Ann Surg Oncol 2004;11:71-7. [Crossref] [PubMed]
- Shuch B, Amin A, Armstrong AJ, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol 2015;67:85-97. [Crossref] [PubMed]
- Durinck S, Stawiski EW, Pavia-Jimenez A, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet 2015;47:13-21. [Crossref] [PubMed]
- Burke HB. Outcome prediction and the future of the TNM staging system. J Natl Cancer Inst 2004;96:1408-9. [Crossref] [PubMed]
- Park YH, Lee SJ, Cho EY, et al. Clinical relevance of TNM staging system according to breast cancer subtypes. Ann Oncol 2019;30:2011. [Crossref] [PubMed]
- Yalaza M, Inan A, Bozer M. Male Breast Cancer. J Breast Health 2016;12:1-8. [Crossref] [PubMed]
- Sun F, Ma K, Yang X, et al. A nomogram to predict prognosis after surgery in early stage non-small cell lung cancer in elderly patients. Int J Surg 2017;42:11-6. [Crossref] [PubMed]
- Wang Y, Li J, Xia Y, et al. Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin Oncol 2013;31:1188-95. [Crossref] [PubMed]
- Kawai K, Sunami E, Yamaguchi H, et al. Nomograms for colorectal cancer: A systematic review. World J Gastroenterol 2015;21:11877-86. [Crossref] [PubMed]
- Shariat SF, Karakiewicz PI, Suardi N, et al. Comparison of nomograms with other methods for predicting outcomes in prostate cancer: a critical analysis of the literature. Clin Cancer Res 2008;14:4400-7. [Crossref] [PubMed]
- Deng H, Qi X, Zhang Y, et al. Diagnostic accuracy of contrast-enhanced computed tomography for esophageal varices in liver cirrhosis: a retrospective observational study. J Evid Based Med 2017;10:46-52. [Crossref] [PubMed]
- Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982;143:29-36. [Crossref] [PubMed]
- Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15:361-87. [Crossref] [PubMed]
- Valentini V, van Stiphout RG, Lammering G, et al. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol 2011;29:3163-72. [Crossref] [PubMed]
- Montironi R, Cheng L, Scarpelli M, et al. Pathology and Genetics: Tumours of the Urinary System and Male Genital System: Clinical Implications of the 4th Edition of the WHO Classification and Beyond. Eur Urol 2016;70:120-3.
- Srigley JR, Delahunt B, Eble JN, et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am J Surg Pathol 2013;37:1469-89. [Crossref] [PubMed]
- Morabito RA, Talug C, Zaslau S, et al. Asymptomatic advanced pediatric papillary renal cell carcinoma presenting as a pulmonary embolus. Urology 2010;76:153-5. [Crossref] [PubMed]
- Nouh MA, Kuroda N, Yamashita M, et al. Renal cell carcinoma in patients with end-stage renal disease: relationship between histological type and duration of dialysis. BJU Int 2010;105:620-7. [Crossref] [PubMed]
- Chai X, Sun MY, Jia HY, et al. A prognostic nomogram for overall survival in male breast cancer with histology of infiltrating duct carcinoma after surgery. PeerJ 2019;7:e7837. [Crossref] [PubMed]
- Koh Y, Lim HY, Ahn JH, et al. Phase II trial of everolimus for the treatment of nonclear-cell renal cell carcinoma. Ann Oncol 2013;24:1026-31. [Crossref] [PubMed]
- Choueiri TK, Vaishampayan U, Rosenberg JE, et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol 2013;31:181-6. [Crossref] [PubMed]
- Escudier B, Bellmunt J, Negrier S, et al. Phase III trial of bevacizumab plus interferon alfa-2a in patients with metastatic renal cell carcinoma (AVOREN): final analysis of overall survival. J Clin Oncol 2010;28:2144-50. [Crossref] [PubMed]
- Wen J, Ye F, He X, et al. Development and validation of a prognostic nomogram based on the log odds of positive lymph nodes (LODDS) for breast cancer. Oncotarget 2016;7:21046-53. [Crossref] [PubMed]
- Xu XL, Cheng H, Tang MS, et al. A novel nomogram based on LODDS to predict the prognosis of epithelial ovarian cancer. Oncotarget 2017;8:8120-30. [Crossref] [PubMed]
- Wang C, Yang C, Wang W, et al. A Prognostic Nomogram for Cervical Cancer after Surgery from SEER Database. J Cancer 2018;9:3923-8. [Crossref] [PubMed]
- Bhindi B, Wallis CJD, Boorjian SA, et al. The role of lymph node dissection in the management of renal cell carcinoma: a systematic review and meta-analysis. BJU Int 2018;121:684-98. [Crossref] [PubMed]
- Ljungberg B, Albiges L, Abu-Ghanem Y, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur Urol 2019;75:799-810. [Crossref] [PubMed]
- Motzer RJ, Jonasch E, Agarwal N, et al. Kidney Cancer, Version 2.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:804-34. [Crossref] [PubMed]