
A novel cell-free single-molecule unique primer extension resequencing (cf-SUPER) technology for bladder cancer non-invasive detection in urine
Introduction
Bladder cancer is one of the most common human malignancies, ranking 8th in incidence and 13th in cancer-related deaths worldwide. In China, the morbidity and mortality of bladder cancer rank first in the malignant urinary tumors, bringing a severe threat to human health and a significant economic burden to families and the country (1-4). Clinically, bladder cancer can be divided into non-muscle invasive cancer and muscle-invasive cancer, and their biological characteristics and treatment strategies are significantly different. Non-muscle invasive bladder cancer, also known as superficial bladder cancer, has a relatively good prognosis. Muscle invasive bladder cancer has a high degree of malignancy and a poor prognosis, and its 5-year survival rate is only 50% to 60% (2,4).
Recently, the leading role of cystoscopy in the diagnosis and recurrence monitoring of bladder cancer due to their low specificity and sensitivity. Cystoscopy is an operator-dependent invasive procedure, and patients suffer from significant pain and high medical expenses. Moreover, the sensitivity of cystoscopy for the diagnosis of bladder cancer in situ is not high enough, and about 10% of the lesions are missed. Besides, cystoscopy may also cause complications such as urinary tract infections and bleeding (5,6). Another commonly used method is urine exfoliated cytology test. The technique has high sensitivity in G3 and high-grade tumors (84%), but low sensitivity in G1and low-tumors (16%) (5,7). Therefore, the search for novel biomarkers with high sensitivity and specificity for early detection of bladder cancer is urgently needed (8).
The discovery of cell-free DNA (cfDNA) provides a theoretical basis for the development of non-invasive liquid biopsy. In cancer patients, a small fraction of cfDNA is tumor-derived, called circulating tumor DNA (ctDNA). Since somatic mutations refer to the presence of mutations only in tumor DNA but not in normal DNA of the same individual, ctDNA is very specific for individual tumors, and successfully avoids false positives of protein biomarkers (9,10). Thus, a non-invasive test with higher sensitivity and specificity can reduce dependence on cystoscopy or urine exfoliated cytology tests in haematuria patients without bladder cancer.
In the context of precision medicine, liquid biopsy has gradually shown its advantages in clinical research and application because of its non-invasive, easy-to-obtain specimens, and dynamic monitoring of tumor gene profiles (11). With the rapid development of next-generation sequencing (NGS) technology, more and more tumor mutation information can be identified. Based on the NGS platform and liquid biopsy technology, we can detect point mutations of cfDNA in urine, and develop novel biomarkers for early detection and recurrence monitoring of bladder cancer (11,12).
In our study, we aimed to use the self-developed cell-free single-molecule unique primer extension resequencing (cf-SUPER) technology to detect point mutations of cfDNA in the urine of haematuria patients, providing a read-out of presence or absence of bladder cancer.
Methods
Clinical sample collection
This study was approved by the Ethics Committee of Xiangya Hospital of Central South University, Changsha, China (Registration ID: 202001001) and Chinese Clinical Trial Registry (Online Registration ID ChiCTR2000029980). All informed consent has been obtained from the individuals involved in this study. Firstly, we collected 29 paired tumor tissue samples and hematuria urine samples from 29 bladder cancer patients that have been confirmed by cystoscopy and pathological biopsy. These 29 patients included 25 males and 4 females, from 45 to 89 years old, with a mean of 62. Among these 29 patients, 24 were at T1 stage, 3 at T2 stage, and 2 at T3 stage. Then, a total of 100 hematuria patients were involved in our study, including 53 bladder cancer-negative patients and 47 bladder cancer-positive patients that have been confirmed by cystoscopy and pathological biopsy. These 53 hematuria patients without bladder cancer included 43 males and 10 females, from 24 to 68 years old, with mean age of 55. The 47 hematuria patients with bladder cancer included 40 males and 7 females, from 16 to 84 years old, with mean of 63. Moreover, 31 bladder cancer patients were at T1 stage, 11 at T2 stage and 5 at T3 stage. We collected 20 mL (2 tubes for 10 mL) morning urine from each of these hematuria patients. The fresh bladder cancer tissue (about 0.2 cm × 0.2 cm) was obtained during operation, which was washed by normal saline. The fresh tissue was then put into a special 1.5ml frozen tubes containing DNA preserved reagent. The urine and tissue samples need to send to Yearth Biotechnology Co. Ltd the same day.
Sample processing
The urine sample was centrifuged 1,600 ×g for 10 min at 4 °C, urine supernatant was carefully removed into some new vacant 2 mL tubes. The urine supernatant was then centrifuged 12,000 ×g for 10 min at room temperature, and the urine supernatant was removed into new vacant 2 mL tubes again, the precipitation was discarded. The collected urine supernatant was store at −80 °C until DNA isolation. The fresh bladder cancer tissue in 1.5 mL frozen tubes was store at −80 °C until DNA isolation.
DNA extract
The Magnetic Serum/Plasma DNA Maxi Kit (cat DP710-02, Tiangen Biotechnology, China) was used to extract urinary cfDNA from collected urine supernatant according to the manufacture’s instruction. The Blood/Cell/Tissue Genomic DNA Extraction Kit (cat DP304, Tiangen Biotechnology, China) was used to extract DNA from the collected tumor tissues or urine supernatant according to the manufacture’s instruction. The DNA was stored at −80 °C until using. The Multiplex I cfDNA Reference Standard Set (HorizonDx), including with 8 mutations at 5%, 1%, and 0.1% allelic frequencies were used to perform the cf-SUPER study.
Uriprier panel design and cf-SUPER sequencing
According to the data from literature and the Catalogue of Somatic Mutations in Cancer (COSMIC) database, we selected 22 genes tightly associated with bladder cancer to build the panel (Table 1). A total of 740 hotspot mutations in these 22 genes were used to design the primers. A primer pool of 149 primer pairs with 100% predicted coverage of target regions were generated. Specific forward oligos intergrade the unique molecular identifiers (UMI) and sample index sequences, specific backward primers, and universal primers were synthesis by IDT. Library construction was performed through an optimized protocol using KAPA HyperPlus Kit (Kapabiosystems). In briefly, 50 ng genomic DNA from each sample was fragmented and ligased to forward oligos with UMI. After purification using two rounds, 1.2× AMPure XP beads (Beckman), the ligased-product as an input to perform a PCR enrichment with specific backward primers and universal primers. The PCR reaction constructed by 2× Glod 360 Master MIX (Applied Biosystems), 13 µL ligased-product, and 400 mM primer-pairs. The enriched library was purified for 1× AMPure beads and then combined with 400 mM P5/P7 sequencing primers and 25 µL HiFi Master Mix (Kapabiosystems). The DNA library was purified again using 1× AMPure beads. The final library was quantified by ABI 7500 (Applied Biosystems) and sequenced on a NextSeq 500 system (Illumina) with all samples run as paired-end 150 bp reads.
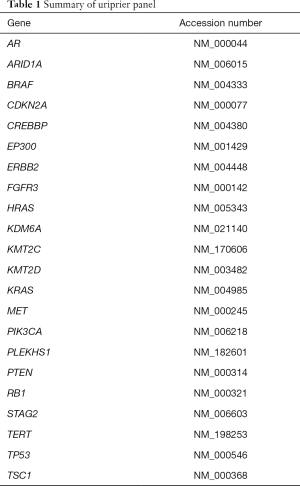
Full table
Sequence data processing and Statistical analysis
The raw data were processed in four steps. First, all reads, including index sequences, the UMI, and adapters were identified and removed. The index sequences and UMI were appended to the read identifier for the next analysis. Second, the trimmed reads were mapped to the human hg19 reference genome using Burrows-Wheeler Aligner (BWA-MEM v0.7.17), only primary aligned reads were applied to the next step. Third, to get the final consensus sequence, the reads with the same UID or in the same loci were cluster. The reads were filtered if the numbers of reads in the same cluster less than 4. The index sequences and UMI were removed from filtered read identifier. Fourth, the variant from the filtered reads was called using Genome Analysis Toolkit (GATK v3.8) and Picard, and then annotated using ANNOVAR. All statistical analysis was processed using GNU R.
Results
Overview and performance of cf-SUPER
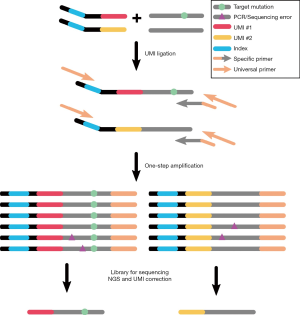
We have developed a protocol integrated UMI and single primer extension (SPE) chemistry, which can detect very low-frequency mutations (Figure 1). Briefly, template DNA is enzymatically fragmented and end-repaired, an adapter containing UMI, index sequences, and a 5’ universal sequence is ligated to the 5’end of DNA fragments. Then PCR cycles have conducted a pool of single primers that contained a specific sequence and a 3’ universal sequence, 5’ universal oligos and Illumina adapter primer. Finally, universal P5/P7 adapter primers from Illumina are used to amplify the sequencing library.
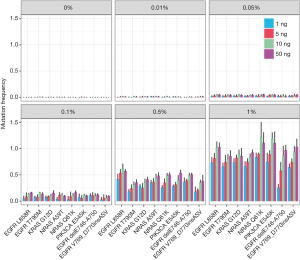
To evaluate the performance of our cf-SUPER method, the Multiplex I cfDNA Reference Standard Set was used to detecting the variant mutations. We found that the cf-SUPER method can accurately detect mutations with allele fractions even low as 0.01% and the DNA input as low as 1 ng (Figure 2).
Consistency evaluation
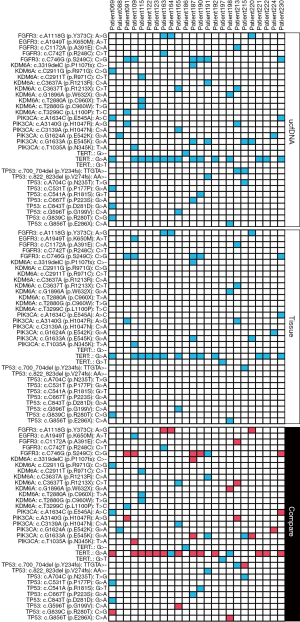
The consistency of mutation profiles between urinary cfDNA, tissue and clinical pathological diagnose was evaluated. In group of urinary cfDNA, at least a same mutation was identified with tissue which was considered consistency, and at least one effective mutation (mutational frequency >0.5%) was tested which was judged as positive case. For the initial 29 paired samples, the mutation tested in urinary cfDNA was 24 cases which were consistent with tissues, 26 cases which were consistent with clinical pathological diagnose, the consistent rate was 82.76%, 89.66%, respectively.
Uriprier panel performance
The uriprier panel was designed to cover in 22 genes. Most of these genes were found in both tumor and urine for the entire cohort (Figure 3). Almost all mutations found in the tumor could be detected from urine samples as well. Besides that, few mutations only detected in urine samples. To evaluate the panel in a large cohort, the samples with bladder cancer from the COSMIC database were used. These genes could be found in more than 80% patients (Figure 4A). Further analysis showed that 470 clinically relevant mutations were adequate enough to cover all of these patients (Figure 4B).
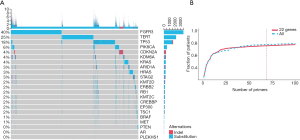
Uriprier panel preclinical evaluation
To evaluate the performance of the uriprier panel, urine samples from 47 patients with hematuria urine were tested using the panel. The panel sensitivity is 97.87% and specificity is 94.34% in these patients (Figure 5A).
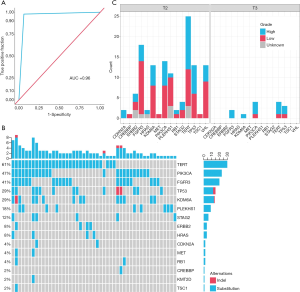
Based on the TNM classification for bladder cancer, we divided two groups of our patient samples. For non-muscle-invasive bladder cancer (Tx to T1) sample, the sensitivity of uriprier panel was 95.83% (23/24), and 100% (23/23) for muscle-invasive bladder cancer (T2-T3) sample (Table 2). The oncoprint (Figure 5B) shows quite similar to the data from the COSMIC. The mutations frequencies are a variety in different stages (Figure 5C).

Full table
Discussion
Urine can be a convenient source of biomarkers for different diseases, mainly for cancer diagnosis, prognosis, treatment monitoring, and prenatal diagnosis (13-15). The advantages of its clinical application are primarily manifested in the non-invasive material and large sample size. Recent studies have shown that urine cell-free DNA (ucfDNA) can provide rich information on somatic cell mutations for studying the genomic characteristics and tumor evolution of patients, making it an important tool for early screening and treatment monitoring of urinary and non-urinary tumors (16,17).
In cancer diagnosis, the occurrence of false or negative results should be minimized, so the high sensitivity and specificity of detection techniques and detection indicators are critical. At present, tumor diagnosis is mainly based on protein biomarkers in peripheral blood, including cancer embryo antigen, prostate-specific antigen (PSA), cancer antigen (CA) 19-9, CA-125 (18,19). Recent studies have shown that urine cfDNA has the potential to become a diagnostic marker for tumors, mainly for bladder and prostate cancer (20).
Concerning the diagnosis of prostate cancer, the GSTP1 gene is most studied, which is usually highly methylated in prostate cancer tissue and can also be detected in serum/plasma and urine cfDNA (21). For non-urological tumors, some somatic cell mutations and methylation changes were detected in the patient’s ucfDNA. For example, A research which detects KRAS mutations in urine/serum/plasma in patients with colorectal cancer found that urine cfDNA provides more abundant mutation information and detects up to 95% of the KRAS mutation ratio than the rate of KRAS mutation detected in the blood cfDNA (35–40%) (22).
For bladder cancer, ucfDNA seems to provide more information than circulating cfDNA. The researcher detected a variety of gene modifications in ucfDNA, including copy number mutations, heterogeneous deficiencies, and some somatic cell mutations, with a 91% sensitivity for bladder cancer diagnosis and only 61% sensitivity based on urine cell DNA testing (23).
In order to find a more powerful diagnose panel, we first screened the COSMIC database. The bladder cancer-related mutations with frequencies greater than 8% were selected, including TERT (51%), FGFR3 (35%), TP53 (29%), KDM6A (21%), PIK3 CA (16%), ARID1A (16%), STAG2 (13%), CDKN2A (11%), RB1 (11%), KMT2D (11%), CREBBP (10%), HRAS (8%), EP300 (9%), KMT2C (9%) and ERBB2 (8%). According to the literature, another 7 bladder cancer-related genes were added, including MET, TSC1, AR, KRAS, BRAF, PLEKHS1, and PTEN (24-30). According to our research, almost all mutations found in the tumor could be detected from urine samples as well. Theoretically and only the coverage of 22 genes shows a similar detection ratio as all genes from COSMIC database with less than 100 pairs of primers and much high than only use 6 common used genes. In this study, 53 hematuria patients without bladder cancer and 47 patients with pathologically confirmed bladder cancer, including 34 in early-stage (phase I) and 13 in mid-stage (phase II–III) were involved in. The detection from all negative controls and all 13 patients with mid-stage bladder cancer is 100% accuracy. By using a 22-gene targeted sequencing panel, we can increase the sensitivity of bladder cancer screening to 98% and 94% specificity. In future studies, it will be necessary to conduct more samples from different research centers to validate these results. Prospective studies could also indicate the risk of bladder cancer relapsing after surgery with urine ctDNA mutations.
In summary, the preliminary work shows that ucfDNA will be the most promising source of biomarkers for future clinical applications, playing an important role in tumor diagnosis and monitoring.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-19-774
Conflict of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-19-774). YP, LL, LW and GX report a patent ZL201910970970.7 licensed to Hunan Yearth Biotechnological Co., LTD. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Xiangya Hospital of Central South University, Changsha, China (Registration ID: 202001001) and Chinese Clinical Trial Registry (Online Registration ID ChiCTR2000029980). All informed consent has been obtained from the individuals involved in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang X, Zhang Y. Bladder Cancer and Genetic Mutations. Cell Biochem Biophys 2015;73:65-9. [Crossref] [PubMed]
- Skeldon SC, Larry Goldenberg S. Bladder cancer: a portal into men’s health. Urol Oncol 2015;33:40-4. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Cao Z, Peng L, He K, et al. Value of quantitative and qualitative analyses of serum and urine cell-free DNA as diagnostic tools for bladder cancer: a meta-analysis. Expert Rev Anticancer Ther 2019;19:645-53. [Crossref] [PubMed]
- Ghafouri-Fard S, Nekoohesh L, Motevaseli E. Bladder cancer biomarkers: review and update. Asian Pac J Cancer Prev 2014;15:2395-403. [Crossref] [PubMed]
- Schmitz-Drager C, Bonberg N, Pesch B, et al. Replacing cystoscopy by urine markers in the follow-up of patients with low-risk non-muscle-invasive bladder cancer?-An International Bladder Cancer Network project. Urol Oncol 2016;34:452-9. [Crossref] [PubMed]
- Yoshino H, Seki N, Itesako T, et al. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol 2013;10:396-404. [Crossref] [PubMed]
- Riethdorf S, Soave A, Rink M. The current status and clinical value of circulating tumor cells and circulating cell-free tumor DNA in bladder cancer. Transl Androl Urol 2017;6:1090-110. [Crossref] [PubMed]
- Tan MP, Attard G, Huddart RA. Circulating Tumour DNA in Muscle-Invasive Bladder Cancer. Int J Mol Sci 2018;19:2568. [Crossref] [PubMed]
- Huang CC, Du M, Wang L. Bioinformatics Analysis for Circulating Cell-Free DNA in Cancer. Cancers (Basel) 2019;11:805. [Crossref] [PubMed]
- Ponti G, Manfredini M, Tomasi A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit Rev Oncol Hematol 2019;141:36-42. [Crossref] [PubMed]
- Gurioli G. Epigenetic Characterization of Cell-Free DNA. Methods Mol Biol 2019;1909:129-35.
- Choi ES, Al Faruque H, Kim JH, et al. Immunochromatographic assay to detect alpha-tubulin in urine for the diagnosis of kidney injury. J Clin Lab Anal 2020;34:e23015. [Crossref] [PubMed]
- Fici P. Cell-Free DNA in the Liquid Biopsy Context: Role and Differences Between ctDNA and CTC Marker in Cancer Management. Methods Mol Biol 2019;1909:47-73.
- Ceylan C, Doluoglu OG, Yahşi S. A different perspective: Can urine pH be important in the diagnosis of prostate cancer? Urologia 2020;87:19-22. [Crossref] [PubMed]
- Elsharkawi F, Elsabah M, Shabayek M, et al. Urine and Serum Exosomes as Novel Biomarkers in Detection of Bladder Cancer. Asian Pac J Cancer Prev 2019;20:2219-24. [Crossref] [PubMed]
- Nonaka K, Matsuda Y, Kakizaki M, et al. Occult mucin-producing urothelial-type adenocarcinoma of the prostate with elevated serum levels of carcinoembryonic antigen and carbohydrate antigen 19-9: Report of an autopsy-proven case. Urol Case Rep 2018;23:6-9. [Crossref] [PubMed]
- Silsirivanit A. Glycosylation markers in cancer. Adv Clin Chem 2019;89:189-213. [Crossref] [PubMed]
- Avogbe PH, Manel A, Vian E, et al. Urinary TERT promoter mutations as non-invasive biomarkers for the comprehensive detection of urothelial cancer. EBioMedicine 2019;44:431-8. [Crossref] [PubMed]
- Bakavicius A, Daniunaite K, Zukauskaite K, et al. Urinary DNA methylation biomarkers for prediction of prostate cancer upgrading and upstaging. Clin Epigenetics 2019;11:115. [Crossref] [PubMed]
- Su J, Lai J, Yang R, et al. Capecitabine plus bevacizumab versus capecitabine in maintenance treatment for untreated characterised KRAS exon 2 wild-type metastatic colorectal cancer: a retrospective analysis in Chinese postmenopausal women. BMC Gastroenterol 2019;19:17. [Crossref] [PubMed]
- Wood HM, Foster JM, Taylor M, et al. Comparing mutation calls in fixed tumour samples between the affymetrix OncoScan(R) array and PCR based next-generation sequencing. BMC Med Genomics 2017;10:17. [Crossref] [PubMed]
- Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47:D941-7. [Crossref] [PubMed]
- Yamasaki K, Mukai S, Nagai T, et al. Matriptase-Induced Phosphorylation of MET is Significantly Associated with Poor Prognosis in Invasive Bladder Cancer; an Immunohistochemical Analysis. Int J Mol Sci 2018;19:3708. [Crossref] [PubMed]
- Guo Y, Chekaluk Y, Zhang J, et al. TSC1 involvement in bladder cancer: diverse effects and therapeutic implications. J Pathol 2013;230:17-27. [Crossref] [PubMed]
- Fenner A. Bladder cancer: Cisplatin response is modulated by AR activity. Nat Rev Urol 2016;13:437. [PubMed]
- Ouerhani S, Bougatef K, Soltani I, et al. The prevalence and prognostic significance of KRAS mutation in bladder cancer, chronic myeloid leukemia and colorectal cancer. Mol Biol Rep 2013;40:4109-14. [Crossref] [PubMed]
- Dudley JC, Schroers-Martin J, Lazzareschi DV, et al. Detection and Surveillance of Bladder Cancer Using Urine Tumor DNA. Cancer Discov 2019;9:500-9. [Crossref] [PubMed]
- Shan G, Tang T, Xia Y, et al. MEG3 interacted with miR-494 to repress bladder cancer progression through targeting PTEN. J Cell Physiol 2020;235:1120-8. [Crossref] [PubMed]


