Analysis of bacterial community using pyrosequencing in semen from patients with chronic pelvic pain syndrome: a pilot study
Introduction
Chronic prostatitis/chronic pelvic pain syndrome (CP/CPPS) is a common condition affecting about 2–6% of men worldwide (1). It is characterized by voiding symptoms with chronic pelvic pain in the absence of a clear bacterial cause. Experts hypothesize that anatomical, neurological, immunological, or psychological disorders are possible etiological factors underlying this condition (2). Thus, there are multiple and diverse treatment options for CP/CPPS. Although empirical antibacterial treatments are currently recommended for inflammatory CP/CPPS, physicians cannot establish the infection in most of the cases. Identification of bacteria based solely on culture methods has often failed to identify specific bacteria that do not grow in general media (3) and underlying pathogens, such as anaerobic bacteria that are not routinely identified by conventional culture methods (4,5).
The 16S ribosomal RNA (rRNA) sequencing technique represents a simple and effective alternative to microbial culture. Sequencing of bacterial 16S rRNA genes from the human body has revealed microbial communities that play an important role in maintaining health and altered microbial communities are associated with a variety of diseases and conditions, including CP/CPPS (6-8). Pyrosequencing is possibly one of the most widely used techniques for 16S rRNA analysis in medical microbiology (9).
Although prostate infections are usually diagnosed by culturing expressed prostatic secretions (EPS) (5), technical variations employed by different physicians may lead to inconsistent results. However, as semen contains diluted prostatic material, some investigators have suggested that semen analysis may be advantageous for the detection of prostatic pathogens (10). Unfortunately, it has not been actively used due to several limitations, including the difficulty of sample collection, preparation, processing, and quality assurance.
In this pilot study, the microbiota of semen was investigated with pyrosequencing to collect evidence of infection in patients with CP/CPPS. Specifically, the pathophysiology of CP/CPPS is at least partly due to dysbiosis of the prostate microbiome (11). Therefore, the identification of various dominant species in CP/CPPS group can facilitate the etiological analysis of CP/CPPS.
Methods
Patients
This prospective, case-control pilot study was conducted at a single university hospital in Korea. Between October 2014 and August 2015, male patients aged ≥19 years and diagnosed with CP/CPPS who had discomfort or pain in the pelvic region ≥3 months during the previous six months and National Institutes of Health Chronic Prostatitis Symptom Index (NIH-CPSI) scores ≥15 were included in the study as the patient group (12). The patients had not been treated with antibiotics for at least three months and had negative urine culture in the screening test. The NIH-CPSI is a representative tool used to assess the symptoms and quality of life in men with CP (13). We also recruited healthy volunteers through an IRB-approved subject recruitment announcement.
Patients who had the following conditions were excluded from the study: (I) cystitis with evidence of a positive culture within the previous three months before entering the study; (II) the presence of genitourinary cancer; (III) patients who underwent intravesical chemotherapy; and (IV) history of pelvic radiation or systemic chemotherapy. Because a significant number of hematospermia patients are known to be associated with evidence for CP/CPPS, patients with hematospermia were not excluded from this study (14).
A physical examination with vital signs, routine laboratory tests, including serum prostate-specific antigen levels and urine analysis, and trans-rectal ultrasonography were performed to evaluate both the healthy volunteers and patients with CP/CPPS at the beginning of the study.
Sample collection
Before semen sampling, the patients were required to cleanse their hands and glans of the penis thoroughly with boric acid-soaked cotton balls. Semen was collected in a sterile 50 mL plastic bottle by masturbation in the clinic. The semen samples were flash-frozen for storage at −20 °C and then sent to a laboratory for analysis. In the laboratory, the DNA was stored at 4 °C until needed. Whole DNA was purified from the semen and the DNA was amplified by polymerase chain reaction (PCR) using universal bacterial primers. All semen samples were also cultured for bacteria by conventional methods. Pyrosequencing analysis of PCR-amplified DNA was also performed.
Sample culture
The semen samples (10 µL) were plated with the use of a pipette. And samples were diluted 1:10 with sterile saline solution (100 µL), vortexed, and plated on blood and MacConkey’s agar. The media were incubated for 24 to 48 hours in atmosphere supplemented at 37 °C. Positive results were from an estimate of the density of each organism ≥103 CFU/mL (15).
Pretreatment of semen samples and DNA extraction
The semen samples were equilibrated to room temperature and centrifuged at 5,000 ×g for 15 min. The supernatant was discarded and the pellet was re-suspended in 1 cc of phosphate-buffered saline before DNA extraction. DNA was extracted from the samples by using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions (16).
Emulsion-based PCR (emPCR)
The extracted genomic DNA was used as a PCR template. A library was prepared using PCR products according to the Genome Sequencer FLX Plus library preparation manual and quantified with the PicoGreen assay using a Victor 3 machine. emPCR was carried out using the Genome Sequencer FLX Plus emPCR Kit (454 Life Sciences, Branford, USA). 16S universal primers, including 27F (5'-GAGTTTGATCMTGGCTCAG-3') and 518R (5'-WTTACCGCGGCTGCTGG-3') were used to amplify the 16s rRNA genes. After the PCR reaction, the products were purified using AMPure beads (Beckman Coulter, FL, USA) (17).
Next-generation sequencing using Roche 454 GS-FLX Plus
Sequencing runs were performed by Macrogen, Ltd. (Seoul, Korea). The emulsion was chemically disrupted and the beads carrying the amplified DNA library were recovered and washed by filtration. Positive beads were purified using biotinylated primer A, which binds to streptavidin-coated magnetic beads. The DNA library beads were separated from the magnetic beads by melting the double-stranded amplification products, leaving a population of bead-bound single-stranded template DNA fragments. The sequencing primer was then annealed to the amplified single-stranded DNA. Lastly, beads carrying amplified single-stranded DNA were counted with a Particle Counter (Beckman Coulter). Sequencing was performed on a Genome Sequencer FLX Plus (454 Life Sciences, Branford, USA) (17).
Selection of 16S rRNAs and taxonomic assignment
Using the basic local alignment search tool (BLASTN), all the sequence reads were compared with the Silva rRNA database. Sequence reads showing sequence similarity with an E-value less than 0.01 were admitted as partial 16S rRNA sequences. Non-16S rRNA sequence reads constituted less than 1%. The taxonomic assignment of the sequenced reads was carried out using the NCBI Taxonomy Databases. Using the database, the five most similar sequences for each sequence read were identified based on their bit scores and E-values derived from the BLASTN program. The Needleman-Wunsch global alignment algorithm was used to determine the optimum alignment of two sequences along their entire length. Pairwise global alignment was performed on selected candidate hits to determine the best aligned hit. The taxonomy of the sequence with the highest similarity was assigned to the sequence read (17).
Operational taxonomic unit (OTU) analysis of community richness
The CD-HIT-OTU software was used for clustering analysis. The mothur software was used to analyze microbial communities and the Shannon diversity and Simpson index were used for species diversity (17).
Ethics statement
This study was approved by the Institutional Review Board of the Catholic University of Korea (VC14TISI0209).
Results
A total of 17 patients with CP/CPPS and four healthy volunteers were enrolled after providing informed consent. The demographic data and baseline characteristics of the subjects are shown in Table S1. The mean ages of the two groups were 51.91±12.27 and 47.25±5.51 years, respectively. The mean NIH-CPSI score was 27.82±5.51 for the patient group and 1.25±0.96 for the control group.
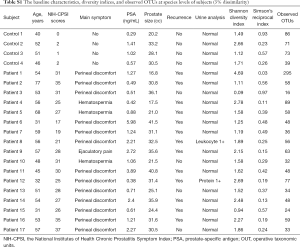
Full table
Bacterial diversity of normal controls and CP/CPPS specimens
None of the semen samples in either group showed colony formation in conventional bacterial cultures. Pyrosequencing of all samples revealed multiple bacterial genera. A total of 232,038 bacterial tag-encoded FLX amplicon pyrosequencing reads were analyzed in this study. With an OTU defined by a similarity cutoff of 97%, which is commonly used to describe the species, 1,343 OTUs were identified.
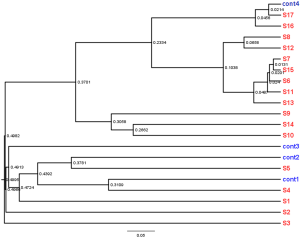
The mean Shannon diversity index score was 1.75±0.66 for the control group and 1.88±0.99 for the patient group, although the mean Simpson index score of the patient group (5.91±0.23) was larger than that of the control group (1.36±0.19) (Table S1, Figure S1). The decreased Shannon and increased Simpson indices suggested decreased diversity in the microbial community in the patient group. No significant difference in bacterial diversity was found between the two groups.
Classification of sequences in normal control and CP/CPPS specimens
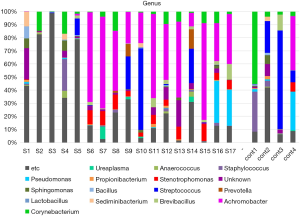
The taxon-based analysis showed an abundance of fastidious bacteria in the semen samples (Figure 1). Corynebacterium, Pseudomonas, Sphingomonas, Staphylococcus, and Streptococcus were frequently detected nonspecifically in both the patient and control groups. However, Achromobacter, Stenotrophomonas, and Brevibacillus were found more frequently in the CP/CPPS patients.
Discussion
The main findings of this prospective, case-control pilot study were: (I) pyrosequencing revealed multiple bacterial genera in both groups, including an abundance of fastidious bacteria, and (II) Achromobacter, Stenotrophomonas, and Brevibacillus were more frequently found in the CP/CPPS patients.
The cause of CP/CPPS is unknown in many cases. It has been attributed to pathophysiological factors, such as pelvic floor dysfunction, neurological disorders, hormonal disturbances, and bacterial infection (18-21). Although these causes appear complex generally, empirical antibacterial treatments are currently recommended for inflammatory CP/CPPS. Specifically, prostatic bacteria can be hidden in calculi or biofilm, increasing resistance (22). These characteristics suggest that prostatic bacteria can cause infections that are difficult to treat (23).
In the field of microbial ecology, molecular biological methods, such as 16S rRNA gene analysis, are generally used without resorting to traditional culture methods. In 1998, Riley et al. investigated diverse and related 16S rRNA-encoding DNA sequences in the prostate of CP patients (24). Sequencing of 36 rDNA clones from 23 rDNA-positive patients confirmed the presence of rDNAs of the Vibrio family, specifically Aeromonas spp., which was previously undetected by culture-based methods. In 2002, Hou et al. used denaturing gradient gel electrophoresis to demonstrate that most of the EPSs derived from CP/CPPS and infertility patients were 16S rRNA gene-positive, unlike in normal men. The studies suggested that the ecological balance of the prostate might play a key role in the maintenance of a healthy reproductive system (25). Recently, Ecker et al. reported specific differences in the microbiome contents of Burkholderia cenocepacia, especially in first-voided urine in CP/CPPS patients compared to controls using next-generation molecular technology in the Multidisciplinary Approach to the Study of Chronic Pelvic Pain (MAPP) Network Epidemiological/Phenotyping Study (26).
Our study also analyzed the differences in the microbiome distribution between CP/CPPS patients and healthy controls. Achromobacter, Sediminibacterium, Stenotrophomonas, and Brevibacillus were found more frequently in patients with CP/CPPS, although Corynebacterium, Streptococcus, Sphingomonas, Pseudomonas, and Staphylococcus were frequently detected in both groups.
Achromobacter species can survive in aqueous environments, such as dialysis fluids, distilled water, deionized water, and humidifiers, which might be a source of contamination in the laboratory (27). Nevertheless, Achromobacter xylosoxidans has been reported as the causative agent in urinary tract infections (28). However, Achromobacter is a common lab contaminant. This point could be a limitation of this study and thus, it is necessary to conduct carefully designed studies with negative controls to confirm its clinical significance. Stenotrophomonas maltophilia has increasingly been recognized as an important cause of nosocomial infections. The types of infections associated with Stenotrophomonas maltophilia include bloodstream infections, pneumonia, and urinary tract infections (29,30). In our study, Stenotrophomonas spp. were detected in 14 patients (82.4%), but the percentage was small. Thus, the role of Stenotrophomonas spp. in CP/CPPS is still unclear.
In addition, Mandar et al. compared the seminal bacterial composition in men with and without CP by sequencing the V6 region of 16S rRNA genes. The findings suggested that the semen of patients with CP contains fewer health-supporting Lactobacilli and exhibits higher species diversity (Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria) than that of healthy controls (31). We also found a relatively low level of Lactobacilli in the CP/CPPS group compared to the control group.
However, our results showed that the effect of Mycoplasma spp. and Ureaplasma spp. on CP/CPPS was less than expected from previous studies, which reported higher frequencies of these species in the semen of patients with CP than in healthy controls (32-34). Corynebacterium species were also associated with CP in previous studies of semen or urine samples (35,36). However, in this study, Corynebacterium species were frequently found in both the CP/CPPS patients and the healthy controls. It was difficult to identify clear differences in any other species between the patients with CP/CPPS and the healthy controls.
The pathophysiology of CP/CPPS has been attributed to dysbiosis of the prostate microbiome, at least partially (11). Therefore, we thought that the identification of various dominant species in our CP/CPPS group other than those reported in previous studies might be of value, although a further carefully designed study including multiple cases is necessary to establish clinical significance.
The limitations of our study are as follows: First, this study was a pilot study comparing bacterial communities using pyrosequencing analysis of semen derived from patients with CP/CPPS and healthy volunteers. Thus, our study had a relatively small sample size and was, therefore, underpowered to reveal any differences. The study merely confirmed the dominant genera. Therefore, a large-sized, well-controlled study is needed to establish the clinical significance of the results. Second, semen samples may contain organisms, which can lead to a misdiagnosis of infection at the prostatic level. The possibility that our samples were contaminated cannot be ruled out, though extreme care was taken during sample procurement and handling to limit contamination.
Conclusions
It was difficult to identify clear differences between CP/CPPS patients and healthy controls in our study due to the limitations associated with the pilot study. However, the detected species were not consistent with previous studies. Therefore, the identification of various dominant species in our CP/CPPS group other than those reported in previous studies might be helpful for future etiological investigations of CP/CPPS.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.02.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Institutional Review Board of the Catholic University of Korea (VC14TISI0209) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Doiron RC, Nickel JC. Management of chronic prostatitis/chronic pelvic pain syndrome. Can Urol Assoc J 2018;12:S161-3. [Crossref] [PubMed]
- Schaeffer AJ. Clinical practice. Chronic prostatitis and the chronic pelvic pain syndrome. N Engl J Med 2006;355:1690-8. [Crossref] [PubMed]
- Srinivasan R, Karaoz U, Volegova M, et al. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS One 2015;10:e0117617. [Crossref] [PubMed]
- Nagy E, Szoke I, Torok L, et al. The role of anaerobic bacteria in prostatitis. Adv Exp Med Biol 2000;485:289-99. [Crossref] [PubMed]
- Domingue GJ Sr, Hellstrom WJ. Prostatitis. Clin Microbiol Rev 1998;11:604-13. [Crossref] [PubMed]
- Shoskes DA, Shahed AR. Detection of bacterial signal by 16S rRNA polymerase chain reaction in expressed prostatic secretions predicts response to antibiotic therapy in men with chronic pelvic pain syndrome. Tech Urol 2000;6:240-2. [PubMed]
- Leskinen MJ, Rantakokko-Jalava K, Manninen R, et al. Negative bacterial polymerase chain reaction (PCR) findings in prostate tissue from patients with symptoms of chronic pelvic pain syndrome (CPPS) and localized prostate cancer. Prostate 2003;55:105-10. [Crossref] [PubMed]
- Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev 2010;74:453-76. [Crossref] [PubMed]
- Siqueira JF Jr, Fouad AF, Rôças IN. Pyrosequencing as a tool for better understanding of human microbiomes. J Oral Microbiol 2012. [Crossref] [PubMed]
- Magri V, Wagenlehner FM, Montanari E, et al. Semen analysis in chronic bacterial prostatitis: diagnostic and therapeutic implications. Asian J Androl 2009;11:461-77. [Crossref] [PubMed]
- Murphy SF, Anker JF, Mazur DJ, et al. Role of gram-positive bacteria in chronic pelvic pain syndrome (CPPS). Prostate 2019;79:160-7. [Crossref] [PubMed]
- Krieger JN, Nyberg L Jr, Nickel JC. NIH consensus definition and classification of prostatitis. Jama 1999;282:236-7. [Crossref] [PubMed]
- Litwin MS, McNaughton-Collins M, Fowler FJ Jr, et al. The National Institutes of Health chronic prostatitis symptom index: development and validation of a new outcome measure. Chronic Prostatitis Collaborative Research Network. J Urol 1999;162:369-75. [Crossref] [PubMed]
- Lee G. Chronic Prostatitis: A Possible Cause of Hematospermia. World J Mens Health 2015;33:103-8. [Crossref] [PubMed]
- Wilson ML, Gaido L. Laboratory diagnosis of urinary tract infections in adult patients. Clin Infect Dis 2004;38:1150-8. [Crossref] [PubMed]
- Kim SJ, Lee DS, Lee SJ. The prevalence and clinical significance of urethritis and cervicitis in asymptomatic people by use of multiplex polymerase chain reaction. Korean J Urol 2011;52:703-8. [Crossref] [PubMed]
- Mir RA, Weppelmann TA, Elzo M, et al. Colonization of Beef Cattle by Shiga Toxin-Producing Escherichia coli during the First Year of Life: A Cohort Study. PLoS One 2016;11:e0148518. [Crossref] [PubMed]
- Lee JC, Muller CH, Rothman I, et al. Prostate biopsy culture findings of men with chronic pelvic pain syndrome do not differ from those of healthy controls. J Urol 2003;169:584-7; discussion 587-8. [Crossref] [PubMed]
- Pontari MA, Ruggieri MR. Mechanisms in prostatitis/chronic pelvic pain syndrome. J Urol 2008;179:S61-7. [Crossref] [PubMed]
- Anderson RU, Wise D, Nathanson BH. Chronic Prostatitis and/or Chronic Pelvic Pain as a Psychoneuromuscular Disorder-A Meta-analysis. Urology 2018;120:23-9. [Crossref] [PubMed]
- Westesson KE, Shoskes DA. Chronic prostatitis/chronic pelvic pain syndrome and pelvic floor spasm: can we diagnose and treat? Curr Urol Rep 2010;11:261-4. [Crossref] [PubMed]
- Nickel JC, Costerton JW. Coagulase-negative staphylococcus in chronic prostatitis. J Urol 1992;147:398-400; discussion 400-1. [Crossref] [PubMed]
- Krieger JN, Riley DE. Bacteria in the chronic prostatitis-chronic pelvic pain syndrome: molecular approaches to critical research questions. J Urol 2002;167:2574-83. [Crossref] [PubMed]
- Riley DE, Berger RE, Miner DC, et al. Diverse and related 16S rRNA-encoding DNA sequences in prostate tissues of men with chronic prostatitis. J Clin Microbiol 1998;36:1646-52. [Crossref] [PubMed]
- Hou DS, Long WM, Shen J, et al. Characterisation of the bacterial community in expressed prostatic secretions from patients with chronic prostatitis/chronic pelvic pain syndrome and infertile men: a preliminary investigation. Asian J Androl 2012;14:566-73. [Crossref] [PubMed]
- Ecker DJ, Sampath R, Massire C, et al. Ibis T5000: a universal biosensor approach for microbiology. Nat Rev Microbiol 2008;6:553-8. [Crossref] [PubMed]
- Gray JS, Birmingham JM, Fenton JI. Got black swimming dots in your cell culture? Identification of Achromobacter as a novel cell culture contaminant. Biologicals 2010;38:273-7. [Crossref] [PubMed]
- Igra-Siegman Y, Chmel H, Cobbs C. Clinical and laboratory characteristics of Achromobacter xylosoxidans infection. J Clin Microbiol 1980;11:141-5. [Crossref] [PubMed]
- Samonis G, Karageorgopoulos DE, Maraki S, et al. Stenotrophomonas maltophilia infections in a general hospital: patient characteristics, antimicrobial susceptibility, and treatment outcome. PLoS One 2012;7:e37375. [Crossref] [PubMed]
- Denton M, Kerr KG. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev 1998;11:57-80. [Crossref] [PubMed]
- Mändar R, Punab M, Korrovits P, et al. Seminal microbiome in men with and without prostatitis. Int J Urol 2017;24:211-6. [Crossref] [PubMed]
- Mändar R, Raukas E, Turk S, et al. Mycoplasmas in semen of chronic prostatitis patients. Scand J Urol Nephrol 2005;39:479-82. [Crossref] [PubMed]
- Gdoura R, Kchaou W, Chaari C, et al. Ureaplasma urealyticum, Ureaplasma parvum, Mycoplasma hominis and Mycoplasma genitalium infections and semen quality of infertile men. BMC Infect Dis 2007;7:129. [Crossref] [PubMed]
- Nassar FA, Abu-Elamreen FH, Shubair ME, et al. Detection of Chlamydia trachomatis and Mycoplasma hominis, genitalium and Ureaplasma urealyticum by polymerase chain reaction in patients with sterile pyuria. Adv Med Sci 2008;53:80-6. [Crossref] [PubMed]
- Tanner MA, Shoskes D, Shahed A, et al. Prevalence of corynebacterial 16S rRNA sequences in patients with bacterial and "nonbacterial" prostatitis. J Clin Microbiol 1999;37:1863-70. [Crossref] [PubMed]
- Türk S, Korrovits P, Punab M, et al. Coryneform bacteria in semen of chronic prostatitis patients. Int J Androl 2007;30:123-8. [Crossref] [PubMed]