
The construction and analysis of competitive endogenous RNA (ceRNA) networks in metastatic renal cell carcinoma: a study based on The Cancer Genome Atlas
Introduction
Renal cell carcinoma (RCC) is a common malignant tumor in the urinary system, and is the tenth leading cancer type in the United States. In 2018 alone, there were 65,340 estimated new cases of RCC and approximately 14,970 deaths due to RCC in the United States (1). The median age at diagnosis of patients with RCC is 64 years old, with 53.5% of RCC patients aged between 55 and 74 years (2). Approximately 15% of patients with RCC present metastatic disease at diagnosis, and 20–40% of patients who are diagnosed with localized RCC will ultimately develop metastatic RCC (mRCC) (1,3,4).
With the development of new treatments in the last ten years, the median overall survival (OS) of patients with mRCC has increased to approximately 30 months, which is a significant improvement compared to the OS of patients 30 years ago. Although the median OS has increased, prognosis is still relatively poor (2). At present, no standard treatment for mRCC exists, and thus combined treatment is adopted (5). Since 2005, molecular-targeted therapies for RCC have rapidly developed, which provides potentially more effective treatment methods (6).
Multiple drugs that target the vascular endothelial growth factor (VEGF) signaling pathway, including sunitinib, pazopanib, and bevacizumab, are approved for treatment of favorable-risk and intermediate-risk mRCC. Several other agents, such as the mammalian target of rapamycin (mTOR) receptor inhibitors and anti-PD-1 immune checkpoint inhibitors, have also shown benefits in response to mRCC therapy (6,7). Thus, a further study on the molecular mechanism of mRCC is needed to identify more effective therapeutic targets and biomarkers for the diagnosis and treatment of mRCC.
In 2011, Salmena et al. first proposed the concept of competing endogenous RNAs (ceRNAs), which deepened our understanding of RNA interactions (8). According to the ceRNA hypothesis, coding RNAs and noncoding RNAs (ncRNAs) may serve as ceRNAs to regulate each other by competing for miRNAs, which share one or more miRNA response elements (MREs) (9). Previous studies on both solid tumors and hematopoietic malignancies have demonstrated that ceRNAs play significant roles in cancer pathogenesis (10). Yin et al. reported that a long-noncoding RNA (lncRNA)-related ceRNA network was involved in clear cell RCC (ccRCC) and its components could serve as independent biomarkers that predict survival in patients with ccRCC (11). However, to the best of our knowledge, the mechanism of ceRNA network in mRCC remains poorly understood.
In this study, we first obtained RNA expression profiles from The Cancer Genome Atlas (TCGA) Database. We then built a differentially expressed lncRNA-miRNA-mRNA ceRNA network for mRCC by comparing, predicting, and integrating differentially expressed RNAs. Lastly, we performed an OS analysis on lncRNAs and mRNAs in the ceRNA network to investigate potential prognostic biomarkers of mRCC. This study aims to provide a new strategy for the prognosis and treatment of mRCC.
Methods
Acquisition of patient information from TCGA database
The TCGA database provides high-throughput data analyses of differentially expressed genes, which include clinical sample information and sequencing data of mRNAs, lncRNAs, and miRNAs. In this study, we used the Data Transfer Tool to download expression data on mRNAs, lncRNAs, and miRNAs from mRCC samples, which contained numerous independent data files that each correspond to one sample. A total of 50 mRCC samples and 74 ccRCC samples were included for further analysis.
Analysis of differentially expressed lncRNAs, miRNAs, and mRNAs
Using the Empirical Analysis of Digital Gene Expression Data package in the software R (edgeR), the downloaded data were normalized and analyzed to identify differentially expressed mRNAs (DEmRNAs), lncRNAs (DElncRNAs), and miRNAs (DEmiRNAs). We then selected the intersection of differentially expressed mRCC lncRNAs, mRNAs, and miRNAs for subsequent analysis. All data from TCGA are publicly available and their use do not require the approval of a local ethics committee.
Construction of ceRNA Network
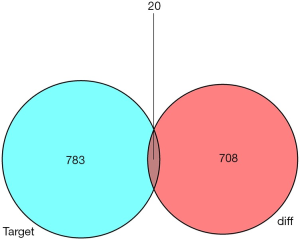
Based on the current ceRNA theory, it is already known that mRNAs can be affected by lncRNAs through a mechanism that involves miRNAs (8). We first used miRcode (http://www.mircode.org/) to predict interactions between lncRNAs and miRNAs based on the differential expression of mRCC miRNA. Secondly, miRanda (http://www.microrna.org/microrna/home.do), miRDB (http://www.mirdb.org/), and TargetScan (http://www.targetscan.org/) were used to predict the miRNA-targeted mRNAs. A Venn diagram was used to improve the reliability of the bioinformatics analysis and to obtain the portion of the target mRNA that overlapped with the differentially expressed mRNAs in mRCC samples, which were further analyzed as DEmRNAs. We then determined the intersection of the differentially expressed lncRNAs and mRNAs, and lastly established the lncRNA/mRNA/miRNA ceRNA network using Cytoscape v3.0.
Functional enrichment analysis
Understanding the biological function and pathway of the genes that are involved in the ceRNA network is highly important. Hence, a Gene Ontology (GO) functional enrichment analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) with a false discovery rate (FDR) of <0.01 as the cutoff value. Further, we conducted a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis using the ClusterProfiler package in the R language software package (adjusted P value is <0.05).
Survival analysis
We used a Cox regression analysis of the survival packages and Kaplan-Meier curves in R to assess the relationship between DElncRNAs, DEmiRNAs, and DEmRNAs expressions. Further, the OS clinical data of patients with mRCC were obtained from TCGA database, which contains information such as gene expression, prognosis, and survival time. A P value of <0.05 is considered statistically significant.
Results
Differentially expressed RNAs in mRCC
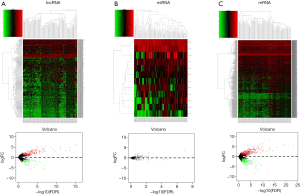
We explored the differential expressed lncRNAs, mRNAs, and miRNAs by comparing the primary renal carcinoma samples with the mRCC samples. An absolute log 2-fold change of >2.0 and a P value <0.01 were considered as cutoff conditions for the screening process. Therefore, a total of 369 DElncRNAs, 12 DEmiRNAs, and 728 DEmRNAs were identified. Among these RNAs, 251 DElncRNAs, 7 DEmiRNAs, and 440 DEmRNAs were upregulated. A cluster analysis of RNA heatmaps and volcano plots is presented in Figure 1.
The ceRNA network in mRCC
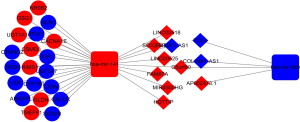
To further study the ceRNA mechanism in mRCC, a ceRNA network was established. We used miRcode to predict interactions between lncRNAs and miRNAs, which resulted in 12 lncRNA-miRNA pairs found. Subsequently, miRDB, miRanda, and TargetScan were used to predict miRNA-targeted mRNAs, resulting in 20 identified target mRNAs. As shown in Figure 2, the 20 target mRNAs, which were part of the 728 previously identified DEmRNAs, were involved in the ceRNA network. Cytoscape software was then used to construct the ceRNA network (Figure 3). A total of 11 lncRNAs, 20 mRNAs, and 2 miRNAs were included in the mRCC ceRNA network.
Functional enrichment analysis of mRNA in the ceRNA network
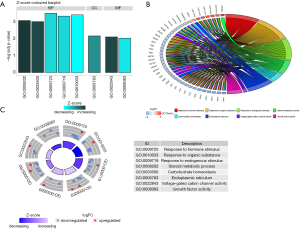
We performed a GO enrichment analysis and KEGG pathway analysis using DAVID and KOBAS (KEGG Orthology Based Annotation System) to further investigate the function of our ceRNA network, the results of which reveal 8 GO terms and 4 KEGG pathways. The GO terms and KEGG pathways of DEmRNAs are presented in Figure 4 and Table 1, respectively.

Full table
As exhibited in Figure 4, DEmRNAs were mainly enriched in processes such as “response to hormone stimulus”, “response to organic substance”, “steroid metabolic process”, “voltage-gated cation channel activity”, and “growth factor activity”. The KEGG pathway analysis reveals that the synthesis and metabolism of hormones and the adipocytokine signaling pathway were involved in the ceRNA network. These results indicate that the ceRNA network may modulate various functions and regulate multiple signaling pathways in mRCC development.
Survival analysis of DElncRNAs and DEmRNAs in mRCC
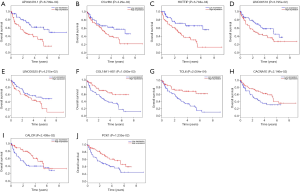
We also used the Kaplan-Meier curve analysis to assess the prognostic values of the identified DElncRNAs and DEmRNAs in mRCC. We found that 7 DElncRNAs were related to mRCC prognosis. Among the 7 DElncRNAs, high expression levels of 5 lncRNAs (AP002478.1, C5orf60, HOTTIP (HOXA transcript at the distal tip), LINC00518, and LINC00525) were associated with poor mRCC prognosis (Figure 5A,B,C,D,E). Nevertheless, high expression levels of lncCOL18A1-AS1 and lncTCL6 were associated with high survival rates (Figure 5F,G). In addition, only 3 mRNAs were associated with mRCC prognosis (Figure 5H,I,J): high expression levels of CACNA1E (calcium channel, voltage-dependent, R-type, α1E subunit), CALCR (calcitonin receptor), and PCK1 (phosphoenolpyruvate carboxykinase 1) were associated with high survival rates.
Discussion
Renal cancer is a common malignant tumor of the urinary system, and its incidence is increasing each year (1,11). More than 400,000 individuals are newly diagnosed with renal cancer annually, while more than 175,000 individuals die from this disease worldwide (12). MRCC is the most lethal form of kidney cancer (12), and thus warrants further investigation on the underlying molecular and pathogenetic mechanisms.
Previous studies have proved that lncRNAs play important roles in tumor progression and metastasis. For instance, one lncRNA, referred to as EGFR-AS1 (EGFR antisense RNA 1), promotes cell growth and metastasis in RCC (13). In prostate cancer, the lncRNA known as SChLAP1 (SWI/SNF complex antagonist associated with prostate cancer 1) may serve as a meaningful biomarker to predict patients with prostate cancer at high risk of lethal progression (14).
According to the ceRNA hypothesis, lncRNAs can act as ceRNAs in their communications with mRNAs by competing for shared MREs and regulating the expression of target genes (8). Research on the role of ceRNA in various tumors has rapidly increased, and the importance of ceRNA for cancer progression is gradually being elucidated (8). For instance, Hong et al. found that a lncRNA, known as HOTAIR (HOX transcript antisense RNA), functioned as a ceRNA for miR-217 to facilitate HIF-1α (hypoxia inducible factor 1 subunit alpha) expression and upregulated AXL levels, which promoted RCC proliferation, migration, and the epithelial-mesenchymal transition (EMT) process (15). Further, lncRNA-ATB promotes cell invasion and metastasis in hepatocellular carcinoma by competitively binding to the miR-200 family with ZEB1 (zinc finger E-box binding homeobox 1) and ZEB2 (zinc finger E-box binding homeobox 2) (16). In gastric cancer, GAPLINC (gastric adenocarcinoma associated, positive CD44 regulator, long intergenic non-coding RNA), which is a lncRNA, can promote cellular proliferation and invasion by acting as a ceRNA of CD44 and competing for miR-211-3p (17). Therefore, we consider that the ceRNA mechanism plays an important and crucial role in the development of tumors.
Consequently, the construction of ceRNA networks is of great significance for further research on the mechanisms of cancer. Furthermore, the construction of ceRNA networks in multiple tumors, such as breast cancer, lung cancer, and colorectal cancer, has been previously reported in scientific literature (18-20). However, few studies on ceRNA in mRCC have been reported. Therefore, in this study, we studied the mechanism of the ceRNA network in mRCC.
In the present study, we analyzed differentially expressed RNAs between mRCC samples and RCC samples and identified 369 DElncRNAs, 12 DEmiRNAs, and 728 DEmRNAs. A ceRNA network of the identified RNAs was then constructed using bioinformatics tools. Further, we found 11 lncRNAs, 2 miRNAs, and 20 mRNAs that were involved in the ceRNA network. We lastly performed GO and KEGG analyses of the mRCC-specific mRNAs.
The GO enrichment results revealed that the functions of differentially expressed genes were enriched in response to hormone stimuli, organic substances, endogenous stimuli, steroid metabolic processes, carbohydrate homeostasis, the endoplasmic reticulum, voltage-gated cation channel activities, and growth factor activities. The KEGG pathway enrichment results indicate that the pathways of steroid hormone biosynthesis, type II diabetes mellitus, xenobiotics metabolism, and adipocytokine signaling are the pathways that are most associated with mRCC.
Tumor cells metastasize when under unfavorable conditions, such as hypoxia, oxidative stress, and inadequate amino acid supply, tend to compromise protein folding in the endoplasmic reticulum (21). Soluble growth factors in the microenvironment play an important role in tumor development, invasion, and metastasis, as well as in the response to targeted anti-tumor therapy (22). The adipokine signaling pathway plays various roles in the initiation, progression, metastasis, and drug response of breast cancer, and can be an effective target for drugs (23). These functions and pathways are closely related to the development, invasion, and metastasis of mRCC. Thus, the ceRNA network plays a vital role in the progression and metastasis of mRCC.
In this study, we further explored the potential mechanism of the intersections among lncRNAs, miRNAs, and mRNAs in mRCC and found that 7 DElncRNAs (AP002478.1, C5orf60, COL18A1-AS1, HOTTIP, LINC00518, LINC00525, and TCL6) and 3 DEmRNAs (CACNA1E, CALCR, and PCK1) were significantly related to OS. Based on the survival analysis, the expression levels of two lncRNAs more specifically COL18A1-AS1 and TCL6 showed positive correlations with survival time. However, the expression levels of the other 5 lncRNAs showed negative correlations with OS. In fact, it has been found that one lncRNA, referred to as AP002478.1, may act as a potential biomarker for patients with gastric cancer caused by Helicobacter pylori (24). Further, the lncRNAs COL18A1-AS1 and TCL6 may play important roles in the pathogenesis of ccRCC and serve as potential biomarkers (25). Importantly, overexpression of the lncRNA Linc00518 in prostate cancer contributes to paclitaxel resistance by sequestering miR-216b-5p (26). Wang et al. found that lncRNA LINC00525 may act as a ceRNA that competes with miR-507 to regulate ELK3 expression in colorectal cancer (27). However, there are no reports on the role of C5orf60, as a lncRNA, in other diseases. HOTTIP, which is also a lncRNA, serves as a ceRNA to play an important role in multiple diseases, including tumors (28). For instance, Wang et al. found that the ceRNA regulatory network of HOTTIP/miR-615/IGF-2 plays a critical role in RCC progression and potentially contributes to improving RCC diagnosis and therapy (28). Another study revealed that HOTTIP regulates HMGB3 expression by acting as a molecular sponge of miR-615-3p in non-small cell lung cancer (NSCLC) cells and thus may be a promising therapeutic strategy for NSCLC treatment (29). Moreover, findings by Mao et al. suggest that the ceRNA regulatory network of HOTTIP/miR-455-3p/CCL3 plays an important role in osteoarthritis pathogenesis (30). Thus, we speculate that these lncRNAs may play crucial roles in mRCC pathogenesis and serve as potential biomarkers.
The expression of HOTTIP was upregulated in mRCC samples compared to control samples. We found that high HOTTIP expression was associated with poor prognosis in mRCC. According to the ceRNA network that we have constructed, the lncRNA HOTTIP may compete with mir-141 to mediate PCK1 or CALCR mRNA expression, which our analysis shows is positively associated with OS. However, our analysis shows that both PCK1 and CALCR expression are downregulated in mRCC; thus, there may be other factors regulating PCK1 and CALCR expression. Moreover, we observed that high CACNA1E expression was associated with positive prognosis in mRCC. Although seven lncRNAs and three mRNAs that were closely related to the diagnosis and prognosis of mRCC were identified using bioinformatics analysis, the complex molecular regulation mechanisms among these RNAs require further research.
In conclusion, this study produced a comprehensive analysis of lncRNA, miRNA, and mRNA expression profiles and clinical data of patients with mRCC in TCGA database. We used bioinformatics to analyze and construct a ceRNA network of mRCC-specific lncRNAs, miRNAs, and mRNAs based on the ceRNA hypothesis. We then identified several lncRNAs and mRNAs that are closely associated with OS, which may therefore hold prognostic significance. This study further demonstrates the contribution of the ceRNA mechanism in the pathogenesis of mRCC. Although this study evidently has clinical implications, it still has some limitations. Overall, these findings deepen the understanding of mRCC pathogenesis and provide potential therapeutic targets and biomarkers.
Acknowledgments
We are grateful for having had free access to the TCGA databases.
Funding: This work was funded and supported by the National Natural Science Foundation of China (No. 81871151) and the Class A of Major Medical Talents in the Jiangsu Province (No. ZDRCA2016009).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau.2020.02.17). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Adashek JJ, Salgia MM, Posadas EM, et al. Role of Biomarkers in Prediction of Response to Therapeutics in Metastatic Renal-Cell Carcinoma. Clin Genitourin Cancer 2019;17:e454-60. [Crossref] [PubMed]
- Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ 2014;349:g4797. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43-9. [Crossref] [PubMed]
- Escudier B. Combination Therapy as First-Line Treatment in Metastatic Renal-Cell Carcinoma. N Engl J Med 2019;380:1176-8. [Crossref] [PubMed]
- Méjean A, Ravaud A, Thezenas S, et al. Sunitinib Alone or after Nephrectomy in Metastatic Renal-Cell Carcinoma. N Engl J Med 2018;379:417-27. [Crossref] [PubMed]
- Osawa T, Takeuchi A, Kojima T, et al. Overview of current and future systemic therapy for metastatic renal cell carcinoma. Jpn J Clin Oncol 2019;49:395-403. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Cheng Y, Geng L, Wang K, et al. Long Noncoding RNA Expression Signatures of Colon Cancer Based on the ceRNA Network and Their Prognostic Value. Dis Markers 2019;2019:7636757. [Crossref] [PubMed]
- Wang Y, Hou J, He D, et al. The Emerging Function and Mechanism of ceRNAs in Cancer. Trends Genet 2016;32:211-24. [Crossref] [PubMed]
- Yin H, Wang X, Zhang X, et al. Integrated analysis of long noncoding RNA associated-competing endogenous RNA as prognostic biomarkers in clear cell renal carcinoma. Cancer Sci 2018;109:3336-49. [Crossref] [PubMed]
- Kotecha RR, Motzer RJ, Voss MH. Towards individualized therapy for metastatic renal cell carcinoma. Nat Rev Clin Oncol 2019;16:621-33. Erratum in: Nat Rev Clin Oncol 2019 Apr 25. [Crossref] [PubMed]
- Wang A, Bao Y, Wu Z, et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis 2019;10:154. [Crossref] [PubMed]
- Mehra R, Udager AM, Ahearn TU, et al. Overexpression of the Long Non-coding RNA SChLAP1 Independently Predicts Lethal Prostate Cancer. Eur Urol 2016;70:549-52. [Crossref] [PubMed]
- Hong Q, Li O, Zheng W, et al. LncRNA HOTAIR regulates HIF-1α/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis 2017;8:e2772. [Crossref] [PubMed]
- Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014;25:666-81. [Crossref] [PubMed]
- Hu Y, Wang J, Qian J, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res 2014;74:6890-902. [Crossref] [PubMed]
- Gao C, Li H, Zhuang J, et al. The construction and analysis of ceRNA networks in invasive breast cancer: a study based on The Cancer Genome Atlas. Cancer Manag Res 2018;11:1-11. [Crossref] [PubMed]
- Li X, Li B, Ran P, Wang L. Identification of ceRNA network based on a RNA-seq shows prognostic lncRNA biomarkers in human lung adenocarcinoma. Oncol Lett 2018;16:5697-708. [PubMed]
- Liang Y, Zhang C, Ma MH, et al. Identification and prediction of novel non-coding and coding RNA-associated competing endogenous RNA networks in colorectal cancer. World J Gastroenterol 2018;24:5259-70. [Crossref] [PubMed]
- Oakes SA. Endoplasmic reticulum proteostasis: a key checkpoint in cancer. Am J Physiol Cell Physiol 2017;312:C93-102. [Crossref] [PubMed]
- Niepel M, Hafner M, Pace EA, et al. Analysis of growth factor signaling in genetically diverse breast cancer lines. BMC Biol 2014;12:20. [Crossref] [PubMed]
- Cha YJ, Koo JS. Adipokines as therapeutic targets in breast cancer treatment. Expert Opin Ther Targets 2018;22:941-53. [Crossref] [PubMed]
- Liu Y, Zhu J, Ma X, et al. ceRNA network construction and comparison of gastric cancer with or without Helicobacter pylori infection. J Cell Physiol 2019;234:7128-40. [Crossref] [PubMed]
- Yang K, Lu XF, Luo PC, et al. Identification of Six Potentially Long Noncoding RNAs as Biomarkers Involved Competitive Endogenous RNA in Clear Cell Renal Cell Carcinoma. Biomed Res Int 2018;2018:9303486. [Crossref] [PubMed]
- He J, Sun M, Geng H, et al. Long non-coding RNA Linc00518 promotes paclitaxel resistance of the human prostate cancer by sequestering miR-216b-5p. Biol Cell 2019;111:39-50. [Crossref] [PubMed]
- Wang S, Li J, Yang X. Long Non-Coding RNA LINC00525 Promotes the Stemness and Chemoresistance of Colorectal Cancer by Targeting miR-507/ELK3 Axis. Int J Stem Cells 2019;12:347-59. [Crossref] [PubMed]
- Wang Q, Wu G, Zhang Z, et al. Long non-coding RNA HOTTIP promotes renal cell carcinoma progression through the regulation of the miR-615/IGF-2 pathway. Int J Oncol 2018;53:2278-88. [PubMed]
- Shi J, Wang H, Feng W, et al. Long non-coding RNA HOTTIP promotes hypoxia-induced glycolysis through targeting miR-615-3p/HMGB3 axis in non-small cell lung cancer cells. Eur J Pharmacol 2019;862:172615. [Crossref] [PubMed]
- Mao G, Kang Y, Lin R, et al. Long Non-coding RNA HOTTIP Promotes CCL3 Expression and Induces Cartilage Degradation by Sponging miR-455-3p. Front Cell Dev Biol 2019;7:161. [Crossref] [PubMed]


