A new era in the detection of urothelial carcinoma by sequencing cell-free DNA
Bladder cancer is a common malignancy with approximately 430,000 new cases diagnosed and 165,000 deaths recorded globally in 2012 (1). Histologically, 90% of bladder cancers originate from the urothelium. Urothelial bladder carcinoma (UBC) is the most common malignancy of the urinary tract. Approximately 25% of UBCs are classified as muscle-invasive bladder cancer (MIBC), which usually requires radical cystectomy, lymph node dissection, and/or perioperative platinum-based chemotherapy. Despite aggressive therapy, a certain number of MIBC patients will experience disease recurrence and die of cancer. Patients with pT2 tumors have relatively good 5-year disease specific survival (DSS), ranging from 70% to 81%. However, patients with ≥ pT3 tumors have a significantly worse prognosis, with 5-year DSS ranging from 40% to 52% (2-5). Pathological node-positivity is also associated with a poor prognosis, with 5-year DSS ranging from 21% to 35% (3,6). For locally advanced MIBC, neoadjuvant platinum-based chemotherapy has been used to improve these unsatisfactory outcomes since the 1980s. Several randomized controlled trials have been performed to define the usefulness of neoadjuvant chemotherapy (7,8); however, platinum-based chemotherapy in the neoadjuvant setting for patients with locally advanced MIBC is still associated with several issues. First, CT or MRI often result in the overdiagnosis or underdiagnosis, with a staging accuracy of only 70% in clinical setting (9). Thus, it is difficult for physicians to evaluate the response to systemic chemotherapy based on radiographic imaging alone. Second, neoadjuvant chemotherapy could delay radical cystectomy in patients who do not respond to chemotherapy and thus experience cancer progression during chemotherapy. A reliable biomarker is necessary for physicians to decide whether to perform neoadjuvant chemotherapy, whether to start immune checkpoint inhibitor, and when to perform radical surgery. There have been several studies about biomarkers to detect disease recurrence, to monitor the efficacy of systemic chemotherapy, or to predict drug response (10-13); however, few biomarkers have shown potential, and there is insufficient evidence to support their routine clinical use.
Recent technological advances, including next-generation sequencing and droplet digital PCR have led genomics, informatics and health information technology to accelerate biomedical discoveries to establish genome-based personalized treatment for many patients. Conventional sampling methods (e.g., needle biopsy) are associated with several problems, including procedural invasiveness, difficulty in obtaining sufficient material of adequate quality, and sampling biases as a result of genetic heterogeneity. Thus, novel diagnostic and molecular biomarkers are needed to help in decision-making in relation to treatment strategies for cancer patients in the clinical setting.
Liquid biopsy technology targeting body fluids, including blood and urine, can offer potential alternatives that overcome the problems associated with conventional biopsy. Cell-free tumor DNA (ctDNA) is shed into the urine or circulation along with DNA from cancer cells. Even though ctDNA constitutes a small fraction of the total DNA, ctDNA is thought to be a promising biomarker. The analysis of ctDNA can provide various types of useful information: including a genomic analysis (without the need for conventional needle biopsy) (14,15), fragment length (16), quantification of the tumor burden (14), and information about the transition of the tumor burden and clonal evolution by serial sampling (17). There are several reports about the clinical utility of plasma ctDNA in selected cancer types. EGFR mutations, which can be detected in the serum or plasma of patients with lung cancer, predict a response to molecular targeted therapies. Thus, the Food and Drug administration approved PCR-based EGFR testing of plasma for patients undergoing first-line treatment for non-small cell lung cancer (18).
In the article entitled “Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficiency by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma”, published in the Journal of Clinical Oncology 2019, Christensen et al. investigated the effectiveness of the assessment of circulating tumor DNA as a useful prognostic biomarker, enabling disease monitoring, early relapse detection, and the potential for earlier clinical intervention in bladder cancer (19). Furthermore, they showed that the ctDNA dynamics during treatment is a predictor of the response to chemotherapy and patient outcome. They performed whole-exome sequencing (WES) of tumor and matched germline DNA from 68 MIBC patients undergoing neoadjuvant chemotherapy, in which they identified 488 mutations (range, 11–3,536 mutations). In reference to the primary tumor WES analysis, they selected 16 highly ranked somatic mutations for unique patient-specific ctDNA assays. Plasma cell-free DNA (cfDNA) was analyzed by ultra-deep multiplex polymerase chain reaction-based next-generation sequencing coverage of 105,000× (error rate: transitions, 0.0063%; transversions, 0.0033%). Among 68 patients, 30 ctDNA-positive cases were detected (44%). They showed that presence of ctDNA at each time point (before chemotherapy, after chemotherapy, and after cystectomy) was strongly associated with recurrence-free survival and overall survival. They observed overall and 12-month recurrence rates among patients whose plasma was ctDNA-positive before chemotherapy of 46% (11 of 24 patients) and 42% (10 of 24 patients), respectively. Whereas only 1 of 35 patients (3%) who were ctDNA-negative before chemotherapy experienced recurrence at 12 months (P<0.001). None of the 35 (0%) patients died during the observational period (P<0.001). This tendency was also observed in ctDNA collected after chemotherapy and after cystectomy. They also analyzed the potential of serial ctDNA measurement (change in the ctDNA level measured in consecutive samples) for disease surveillance. For patients with metastatic relapse and detectable ctDNA, they reported that a ctDNA assay could detect disease recurrence 96 days earlier than conventional radiographic imaging (P=0.023). To assess the impact of tumor heterogeneity between the primary tumor and metastasis, they compared the WES data of the primary tumor with cfDNA obtained at the time of metastatic recurrence after cystectomy. A high degree of similarity was observed in the mutational status of the primary tumor and the cfDNA. They investigated the utility of serial ctDNA measurements for monitoring the therapeutic response. They reported that 85% of ctDNA-negative patients (35 of 41) showed pathologic downstaging. Patients who were initially ctDNA-positive with subsequent clearance of ctDNA showed a response rate of 53% (9 of 17), while patients without ctDNA clearance did not show any response (0 of 8). Clinical parameters and molecular features of the primary tumor were associated with the treatment response and outcome; however, ctDNA monitoring remained the strongest predictor of the outcome
This was the largest and the most comprehensive report about ctDNA in patients with advanced UBC (18). In the study, they documented three important findings for the analysis of ctDNA in patients with advanced UBC: (I) the absence of ctDNA collected at any time point of treatment serves as a powerful biomarker for recurrence free and overall survival; (II) the ctDNA dynamics during chemotherapy reflect the response to treatment and the patient outcome; and (III) ctDNA identifies disease recurrence earlier than radiographic imaging with high sensitivity and specificity after radical surgery. The ctDNA assay that they have reported would be very useful for evaluating the systemic tumor mutation burden because this assay is based on the sequencing of patient-specific and highly mutated genes, and the copies of ctDNA rose linearly along the time course of tumor progression. In addition, that ctDNA-guided management has great potential in cases that are ctDNA-negative at the diagnosis of MIBC, because the RFS and OS of patients without ctDNA was outstanding. These patients may be eligible for immediate cystectomy without neoadjuvant chemotherapy. On the other hand, there should be efforts to improve the prognosis of patients who are ctDNA-positive before chemotherapy, as they are considered to have a high risk of recurrence. High ctDNA copies may indicate a high tumor burden; thus, neoadjuvant chemotherapy with immune check point inhibitors, not platinum-based therapy, might be a good indication for patients with high ctDNA copy numbers. They selected 16 highly ranked mutation genes for each patient for unique patient-specific ctDNA assay; however, in the clinical setting, a lack of versatility might result in an assay that is complicated, costly, time-consuming. Whether the results from the study could be reproduced with a commercial-based targeted exome sequencing panel, and ctDNA copy numbers can serve as a useful prognostic biomarker is a matter of interest. The study showed the great potential for the clinical application of the analysis of ctDNA from patients with advanced UBC. A genomic analysis and the serial sampling of cfDNA could offer a definitive treatment strategy, and provide useful information about tumor heterogeneity, clonal expansion, and drug resistance.
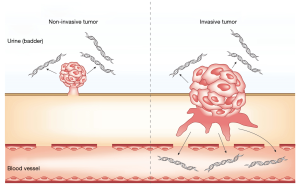
In the setting of cancer screening and the detection of minimal residual disease after radical surgery, it is difficult to maintain analytical sensitivity solely by the detection of mutation-based ctDNA in plasma using current technology. However, urothelial carcinoma is constantly and directly in contact with the urine; thus, some amount of ctDNA and DNA derived from cancer cells is contained in the urine from patients with urothelial carcinoma, and urinary ctDNA and DNA from cancer cells have huge potential in cancer screening and disease monitoring to detect tumor recurrence in urinary tract (Figure 1). Springer et al. reported the utility of UroSEEK, a massive parallel sequencing-based assay, in the detection of UBC and upper tract urothelial carcinoma (UTUC) using urinary pellet DNA (20). UroSEEK consists of three components (UroSeqS, TERTSeqS, and FastTSeqS): the detection of mutations in regions of ten genes (FGFR3, TP53, CDKN2A, ERBB2, HRAS, KRAS, PIK3CA, MET, VHL and MLL); the detection of mutations in the TERT promoter; and the detection of aneuploidy. Among 175 patients, UroSEEK could detect mutations in 145 patients (83%) earlier than macroscopic change. Eich et al. also reported the clinical potential of UroSEEK using tumor DNA from UBC patients. FGFR3 and PIK3CA mutations were more frequently detected in low-grade tumors than high-grade tumors (P<0.0001), while the opposite was true for TP53 mutations (P<0.0001). Significantly higher rates of TP53 and CDKN2A mutations were observed in MIBC than in those with pT1 tumors (21). Besides DNA from cancer cells in urine, Hayashi et al. reported the clinical utility of urinary cfDNA in patients with localized UTUC (22). They studied urinary cfDNA from 153 people and performed droplet digital PCR to detect TERT promoter and FGFR3 hotspot mutations. They detected mutations of TERT C228T, TERT C250T and FGFR3 S249C mutations in 22/56 (39.3%), 4/56 (7.1%), and 9/56 (16.1%) patients, respectively. FGFR3 mutations were only detected in ≤ pT1 tumors. In combination with cytology results, the sensitivity was 78.6%, and the specificity was 96.0%. A mutation analysis of the TERT promoter and FGFR3 in urinary cfDNA has the potential to be a non-invasive diagnostic marker and reliable factor for tumor staging for patients with UTUC. Activating mutations in the promoter of the telomerase reverse transcriptase (TERT) gene led to the increased expression of telomerase and allow some tumors to overcome the end-replication problem and avoid senescence. The mutations in the TERT promoter region were mainly detected in low-self renewal malignancies, including 38% of MIBCs and 32% of NMIBCs (20). Moreover, TERT promoter mutations occur in pre-cancerous lesions, including papillary urothelial neoplasms with low malignant potential (23) and inverted papilloma (24). TERT promoter is therefore the most frequently mutated gene in UBC, and the detection of TERT promoter mutations has the potential to be a reliable biomarker. Although TERT promoter is thought to be a good candidate for a liquid biopsy analysis, the TERT promoter region was not initially identified because of the limited coverage of the promoter region by exome sequencing. Besides, the TERT promoter region is a challenging region to amplify due to its high GC-rich content (>80%), homopolymer runs, and low sequence complexity; thus, the analysis of TERT promoter mutations is running behind the analysis of WES. However, improved PCR techniques, including the use of certain additives to inhibit non-specific reactions, enabled the precise analysis of TERT promoter mutations. We expect further studies on the TERT promoter mutation analysis of cfDNA, which is likely to yield highly valuable information about advanced urothelial carcinoma. In 2018, Robertson et al. investigated five subtypes of UBC and reported the molecular characterization, tumor mutational status and the mRNA expression, and proposed a systemic treatment strategy for advanced UBC based on these subtypes (25). Thus, increased basic research and further clinical trials about precision medicine based on patient genetic information obtained from cfDNA are expected to improve patient outcomes and quality of life.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Mahdavifar N, Ghoncheh M, Pakzad R, et al. Epidemiology, incidence and mortality of bladder cancer and their relationship with the development index in the world. Asian Pac J Cancer Prev 2016;17:381-6. [Crossref] [PubMed]
- Manoharan M, Ayyathurai R, Soloway MS. Radical cystectomy for urothelial carcinoma of the bladder: an analysis of perioperative and survival outcome. BJU Int 2009;104:1227-32. [Crossref] [PubMed]
- Stein JP, Lieskovsky G, Cote R, et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol 2001;19:666-75. [Crossref] [PubMed]
- Ghoneim MA, Abdel-Latif M, el-Mekresh M, et al. Radical cystectomy for carcinoma of the bladder: 2,720 consecutive cases 5 years later. J Urol 2008;180:121-7. [Crossref] [PubMed]
- Madersbacher S, Hochreiter W, Burkhard F, et al. Radical cystectomy for bladder cancer today--a homogeneous series without neoadjuvant therapy. J Clin Oncol 2003;21:690-6. [Crossref] [PubMed]
- Hautmann RE, Gschwend JE, de Petriconi RC, et al. Cystectomy for transitional cell carcinoma of the bladder: results of a surgery only series in the neobladder era. J Urol 2006;176:486-92. [Crossref] [PubMed]
- International Collaboration of Trialists. Neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: a randomised controlled trial. Lancet 1999;354:533-40. [Crossref] [PubMed]
- Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med 2003;349:859-66. [Crossref] [PubMed]
- Sternberg CN, Pansadoro V, Calabrò F, et al. Can patient selection for bladder preservation be based on response to chemotherapy? Cancer 2003;97:1644-52. [Crossref] [PubMed]
- Gómez-Román JJ, Saenz P, Molina M, et al. Fibroblast growth factor receptor 3 is overexpressed in urinary tract carcinomas and modulates the neoplastic cell growth. Clin Cancer Res 2005;11:459-65. [PubMed]
- Hoffmann AC, Wild P, Leicht C, et al. MDR1 and ERCC1 expression predict outcome of patients with locally advanced bladder cancer receiving adjuvant chemotherapy. Neoplasia 2010;12:628-36. [Crossref] [PubMed]
- Ioachim E, Michael MC, Salmas M, et al. Thrombospondin-1 expression in urothelial carcinoma: prognostic significance and association with p53 alterations, tumour angiogenesis and extracellular matrix components. BMC Cancer 2006;6:140. [Crossref] [PubMed]
- Yoshida T, Kates M, Fujita K, et al. Predictive biomarkers for drug response in bladder cancer. Int J Urol. [Epub ahead of print].
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [Crossref] [PubMed]
- Yamamoto Y, Uemura M, Fujita M, et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci 2019;110:617-28. [Crossref] [PubMed]
- Yamamoto Y, Uemura M, Nakano K, et al. Increased level and fragmentation of plasma circulating cell-free DNA are diagnostic and prognostic markers for renal cell carcinoma. Oncotarget 2018;9:20467-75. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Komiya K, Nakashima C, Nakamura T, et al. Current status and problems of T790M detection, a molecular biomarker of acquired resistance to EGFR tyrosine kinase inhibitors, with liquid biopsy and re-biopsy. Anticancer Res 2018;38:3559-66. [Crossref] [PubMed]
- Christensen E, Birkenkamp-Demtröder K, Sethi H, et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J Clin Oncol 2019;37:1547-57. [Crossref] [PubMed]
- Springer SU, Chen CH, Rodriguez Pena MDC, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife 2018;7:e32143. [Crossref] [PubMed]
- Eich ML, Rodriguez Pena MDC, Springer SU, et al. Incidence and distribution of UroSEEK gene panel in a multi-institutional cohort of bladder urothelial carcinoma. Mod Pathol 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Hayashi Y, Fujita K, Matsuzaki K, et al. Diagnostic potential of TERT promoter and FGFR3 mutations in urinary cell-free DNA in upper tract urothelial carcinoma. Cancer Sci 2019;110:1771-9. [Crossref] [PubMed]
- Rodriguez Pena MDC, Tregnago AC, Eich ML, et al. Spectrum of genetic mutations in de novo PUNLMP of the urinary bladder. Virchows Arch 2017;471:761-7. [Crossref] [PubMed]
- Akgul M, MacLennan GT, Cheng L. Distinct mutational landscape of inverted urothelial papilloma. J Pathol 2019;249:3-5. [Crossref] [PubMed]
- Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2018;174:1033. [Crossref] [PubMed]