Head-to-head comparison of prostate cancer risk calculators predicting biopsy outcome
Introduction
Prostate cancer (PCa) is the most prevalent cancer in the western world for men and the second most common cause of death in men worldwide (1). Long-term follow-up from the European Randomised Study of Screening for Prostate Cancer (ERSPC) has shown a significant reduction in PCa specific mortality applying prostate-specific antigen (PSA) based screening (2). This, next to the fact that PSA is a well-developed, easy to implement, and cheap test, made PSA testing the mainstay in the decision for further clinical workup (i.e., prostate biopsy). However, any choice of a PSA cut-off involves a trade-off between sensitivity and specificity. Lowering the PSA cut-off would improve test sensitivity, but also reduce specificity, leading to far more false-positive tests and unnecessary interventions. Additionally, many of the cancers detected may never become clinically evident, thereby leading to overdiagnosis and overtreatment (3,4).
Other relevant pre-biopsy clinical information, next to the serum PSA level, has been incorporated into so-called risk calculators (RCs) to enable a more accurate assessment of a patient’s individual PCa risk (5). A recent systematic review identified 127 unique RCs in the field of PCa. The conclusion was that RCs outperform PSA alone in avoiding unnecessary biopsies, that not all of the RCs have the ability to selectively identify those men at risk of having clinically significant PCa (csPCa) (defined as Gleason score ≥3+4) and that external validation studies and head-to-head comparisons are lacking (6). RCs are part of the European Association of Urology (EAU), European Society for Radiotherapy & Oncology (ESTRO), European Society of Urogenital Radiology (ESUR), and International Society of Geriatric Oncology (SIOG) joint guidelines on PCa screening and early detection (5) but it remains a matter of personal choice whether and/or which RC to use in one’s daily clinical practice.
We aimed to address this lack of information by evaluating the performance (discrimination, calibration, and clinical impact) of the most well-known RCs developed to predict prostate biopsy outcome in a head-to-head comparison.
Methods
Participants
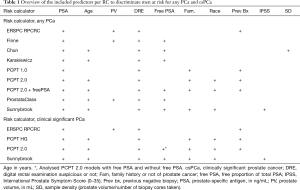
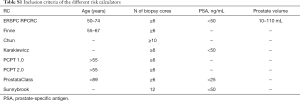
Our study cohort comprises of 8,649 men from ten independent contemporary cohorts (nine in Europe and one in Australia) who underwent a transrectal ultrasound (TRUS)-guided prostate biopsy between January 2007 and November 2015 (Table 1 and Acknowledgements). The cohorts were unrelated to the RCs development. Pre-biopsy clinical data and the pathological biopsy results were obtained from electronic and paper medical charts. Data on age, previous negative biopsy, DRE (digital rectal examination; benign/suspicious), prostate volume (assessed by TRUS in mL), total PSA (tPSA), free PSA (fPSA), and biopsy outcome (PCa detected yes/no and Gleason grade) were collected for all men. Patients were included into the analyses if they met the aggregated criteria of all RCs: age between 50 and 89 years old, PSA <50 ng/mL, prostate volume between 10 and 110 mL (Table S1). Participants in the study underwent a TRUS-guided biopsy according to the standard clinical practice used at each participating site, which was on average 12 cores [interquartile range (IQR): 12–14] per biopsy session in the European cohorts and 30 cores (IQR: 12–30) in the Australian cohort.
Full table
Full table
Statistical analysis
Baseline characteristics of the study cohort are presented as median and interquartile range (M-IQR) or percentage for proportion type features. Missing data was imputed using Multivariate Imputation by Chained Equations (MICE) using only the values from the corresponding cohort and was imputed five times (7). Three parameters were completely missing in certain cohorts: fPSA (Den Bosch, Sydney, Breda), International Prostate Symptom Score (Den Bosch, Porto, Bordeaux, Munster, Paris, Hamburg, Rennes, Milan), and family history of PCa (Den Bosch, Porto, Bordeaux, Munster, Paris, Rennes). These missing values were imputed by using the complete information from the other cohorts in the imputation model. This strategy leads to more precise estimates of the predicted probabilities (8). After imputation, the probabilities were calculated on the basis of the predictions rules from each RC. Individual patient data was compared for men with csPCa and men without PCa using the Mann-Whitney test for continuous variables.
Risk calculators
For this meta-analysis we included seven well known RCs: Chun (9), ERSPC Rotterdam Prostate Cancer Risk Calculator RPCRC (10), Finne (11), Karakiewcz (12), ProstataClass (13), Prostate Cancer Prevention Trial (PCPT) 1.0 and 2.0 +/– free-PSA (14), and Sunnybrook (15). The first six have been externally validated in over five studies (6). All RCs use PSA and DRE as a predictive factor, and the other predictors are displayed in Table 1. The RPCRC for initial and repeat biopsy (16) was adapted to the contemporary Rotterdam clinical setting (17). For each individual patient, the probabilities of having a biopsy detectable PCa and, if applicable, a csPCa, were calculated. The probabilities of the ProstataClass artificial neural network were obtained by sending a blinded database for PCa outcome to the ProstataClass developers (H. Cammann) (13). For uniform comparison between RCs men with a PSA above their advised cut-off for PSA (i.e., >20 ng/mL) were included (157 men (2.2% of the total cohort). Four models were able to separately predict csPCa (RPCRC, PCPT 2.0 +/– freePSA and Sunnybrook).
Comparison of risk calculator models
The predictive accuracy (predicting PCa and csPCa) was quantified using the area under the curve (AUC) for the receiver operator characteristic (ROC) analysis (18). A multivariable meta-analysis was performed to pool the AUCs in predicting any PCa and csPCa. Within-study correlations of the AUCs were estimated using bootstrapping. To analyze statistically significant differences in the models and taking into account the between-study heterogeneity we subsequently estimated the probability that a model has the highest AUC in a subsequent validation study. We simulated 10,000 samples from the posterior distribution to estimate this probability (19). Calibration of the RCs was pooled and explored graphically using calibrations plots. For comparison, the per center sensitivity and specificity of detecting csPCa with applying a PSA cut-off ≥4.0 ng/mL was calculated and graphically displayed.
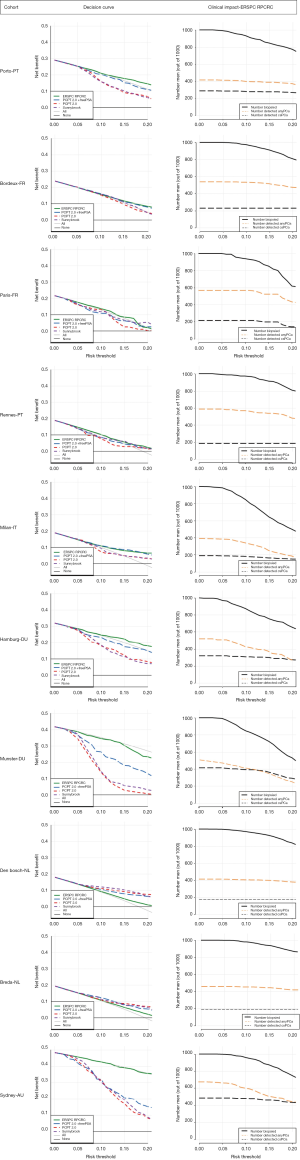
In addition, the clinical impact was assessed with decision curve analysis (DCA) and clinical impact curves. DCA represents the net benefit ratio, which weighs the benefits (detecting cancer) versus the harms (unnecessary biopsy) over a range of thresholds (20). Clinical impact curves show the estimated number who would be declared eligible for biopsy for each risk threshold and show the proportion of those who are cases (overall PCa and csPCa) (21). In addition, to show the potential of multivariate risk stratification when adapting to, for example, one’s own hospital data, we calculated net benefit after calibration of each of the 4 models predicting csPCa using the largest clinical cohort in our series (Den Bosch; N=2,053). Analyses were performed using R statistical package, version 3.3.1, R Foundation for Statistical Computing, Vienna, Austria.
Results
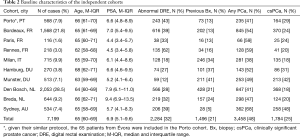
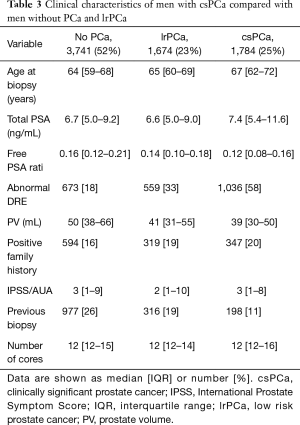
Of the total of 8,649 men 7,119 men (83.2%) were included in analyses. Median age was 65 years old, median PSA 6.9 ng/mL, median prostate volume 45 mL (as evaluated by TRUS) and 1,496 men (21%) underwent a previous biopsy (Table 2). PCa was diagnosed in 3,458 of 7,119 patients (48%) and 1,784 (25%) men had csPCa. PCa patients were older (median age: 65 vs. 64 years for non PCa patients, P<0.001), had smaller prostate glands (41 vs. 50 mL, P<0.001), and higher PSA (6.7 vs. 7.0 ng/mL, P<0.001) (Table 3).
Full table
Full table
In predicting any PCa no particular RC stood out and the pooled area under the ROC-curve (AUC) ranged between 0.64 and 0.72 (Figure 1) with Finne having the highest AUC. Substantial heterogeneity in the AUC was found between the different cohorts (range I2, 66–89%). In predicting csPCa the ERSPC RPCRC had the highest pooled AUC of 0.77 (95% CI: 0.73–0.80; Figure 1). After repeating this comparative analysis 10,000 times the ERSPC RPCRC had the highest probability (89%) of having the highest AUC. The probabilities of having the highest AUC in our study cohort were 6%, 3%, and 2% for PCPT 2.0 + freePSA, PCPT 2.0, and Sunnybrook RCs respectively.
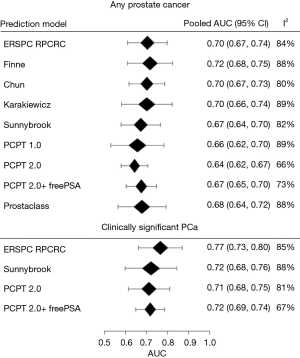
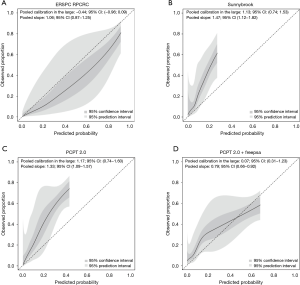
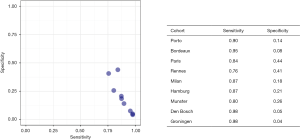
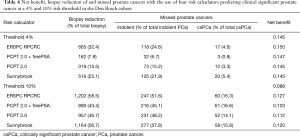
The calibration plots for those RCs predicting csPCa for the pan-European data set (including all cohorts except Sydney, Australia; n=6,665) are displayed in Figure 2. Three models underestimated the probability of csPCa, while the ERSPC RPCRC was more accurate at low probabilities and mainly overestimated at probabilities >10%. Figure S1 displays the decision curves and clinical impact plots per cohort applying the 4 RCs predicting csPCa. Overall the ERSPC RPCRC has the highest net benefit followed by the PCPT 2.0. Clinical impact is negligible to small starting at probabilities >10% for detecting csPCa. Figure S2 displays the differences in sensitivity and specificity of the PSA test (cut-off ≥4.0 ng/mL) per center. Sensitivity and specificity range from 76% to 98% and 4% to 44% respectively. Table 4 shows net benefit when each of the 4 RCs are calibrated. At a 4% threshold for csPCa using the ERSPC RPCRC the number of biopsies can be reduced by 32% while keeping a 95% sensitivity for detecting csPCa. Reduction and sensitivity are 8% and 99% for PCPT2.0 + freePSA, 16% and 97% for PCPT 2.0, and 25% and 95% for Sunnybrook.
Full table
Discussion
In this head-to-head comparison of seven well-known RCs predicting prostate biopsy outcome it is shown that all RCs have a moderate to well discriminatory ability when predicting any PCa (AUCs ranging from 0.64 to 0.72). Those RCs that can selectively predict csPCa show AUCs in the range of 0.71 to 0.77 with small clinical benefit in this pan-European cohort of contemporary daily clinical practice and clinical study data. Adjusting calibration shows the added value of incorporating multivariable risk prediction tools next to clinical expertise in clinical decision making. These results confirm earlier analyses on the use of multivariable prediction tools (6) and considering the substantial harm related to overdiagnosis of low risk PCa (3,22), the use of those RCs that selectively can predict csPCa is recommended.
The balance between benefit and harm of early detection of PCa is still a topic of ongoing debate. Due to the initiation of PSA screening in combination with TRUS guided systematic prostate biopsy the incidence of predominantly low risk PCa increased enormously in the 1990s. This eventually resulted in guidelines recommending no screening at all (23). However, with the available data from longitudinal studies and randomized PCa screening trials we currently have, the knowledge to improve the balance between harm and benefit recommendations have changed to using shared decision making with an individual approach towards how to screen best (24).
While both discrimination and calibration are important statistics to evaluate performance of a prediction tool we must note that discrimination cannot be easily improved while calibration can (6). An example of potentially adjusting calibration to a particular setting is shown in (25) where a model was first tested using part of the available data (calibration phase) where subsequently performance was assessed in the rest of the data (validation cohort). Based on the wide calibration prediction intervals in the current analyses it is advisable to follow such an approach where the aim should be to assess moderate calibration on the basis of center specific retrospective data on prostate biopsy outcome with a minimum of 200 prostate cancers cases (26). Subsequently these center specific adjustments for the calculated probabilities could be incorporated, for example, in the RPCRC. It is in this context important to realize that when using a purely PSA based approach considerable variation in sensitivity and specificity also exists, as was shown in Figure S2, something that is ignored in recommendations on applying a cut-off value to trigger prostate biopsy.
When predicting biopsy outcome, it must also be noted that especially the use of the multiparametric magnetic resonance imaging (mpMRI) in the detection of PCa and csPCa has increased considerably showing very promising results. However, while the mpMRI is advised to be used after a negative TRUS guided systematic prostate biopsy (often solely based on an elevated PSA level), the mpMRI is more and more used before the first biopsy (27). Previous analyses with the RPCRC have however shown that upfront risk stratification on the basis of easy (and cheap) to get relevant pre-biopsy information can avoid half of mpMRIs (17).
This study has some limitations. First, it is a retrospective study design using ten different cohorts from populations with different background risk and different referral patterns (daily clinical practice cohorts with the risk of selective outcome reporting and clinical study cohorts with predefined eligibility criteria) as reflected in the heterogeneity of our results. On the other hand, evaluating performance of these RCs in this pan-European setting can be seen as a strength and support their use in Europe. Second, we mainly used the original RCs which were virtually all developed in the 1990s. This implies that they do not use later developed biomarkers (e.g., PHI, PCA3, the 4K panel). All these biomarkers have shown to have additional predictive value when incorporated into a prediction model and as such might be able to positively influence results (28-30).
Finally, all RCs use as endpoint csPCa based on the original Gleason grading (31). It has been shown that the new Gleason grading system better reflects disease burden (32) as does the inclusion of cribriform growth patterns in the classification of Gleason 7 PCa (16,33).
In conclusion, we performed the first head-to-head comparison of RCs predicting prostate biopsy outcome using a multicenter European and Australian population. No particular RC stood out in the discrimination of men with and without PCa. The ERSPC RPCRC showed highest discrimination when predicting clinically significant PCa. Net benefit in the available clinical cohorts was limited but can be increased by applying a simple calibration step. These outcomes support implementing next to clinical expertise a multivariable risk prediction tool before further workup (e.g., MRI and biopsy) in men suspicious for having a clinically significant PCa.
Acknowledgements
We would like to thank the investigators, researchers and hospitals for providing the required data (listed according to the number of men they enrolled): H. Beerlage, Jeroen Bosch Hospital, ‘s-Hertogenbosch, NL; R. Gaston, T. Piechaud, Clinique St. Augustin, Bordeaux, FR; M. Lazzeri, San Raffaele Hospital-Turro, Milan, IT; S. Roemeling, University Medical Center Groningen: Amphia Hospital Breda, NL; D van der Schoot, Amphia Hospital, Breda, NL; I. Braga, L. Osório, V. Cavadas, A. Fraga, Porto Hospital Center, Porto, PT. E. Carrasquinho, E. Cardoso de Oliveira, Hospital Espírito Santo, Évora, PT; P. Stricker, J. Thompson, P. van Leeuwen, Garvan Institute of Medical Research, University of New South Wales, Sydney, AUS; A. Semjonow, University Hospital Munster, Munster, Germany. C. Stephan, Charite-Universitaetsmedizin, Berlin and Berlin Institute for Urologic Research, Berlin, GE; A. Haese and M. Graefen, Prostate Cancer Center, Martini Clinic, University Hamburg-Eppendorf, Hamburg, GE; S. Vincendeau, Hospital Pontchaillou, Rennes, France. A. Houlgatte, HIA Du Val De Grace, Paris, France; Special thanks to H. Cammann, Universitätsmedizin Berlin, Berlin, DE for calculating the predicted probabilities for ProstataClass ANN.
Footnote
Conflict of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by institutional ethics committees and informed/oral consent was taken from all the patients.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate cancer mortality: results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet 2014;384:2027-35. [Crossref] [PubMed]
- Heijnsdijk EA, Wever EM, Auvinen A, et al. Quality-of-life effects of prostate-specific antigen screening. N Engl J Med 2012;367:595-605. [Crossref] [PubMed]
- Loeb S, Vellekoop A, Ahmed HU, et al. Systematic review of complications of prostate biopsy. Eur Urol 2013;64:876-92. [Crossref] [PubMed]
- Mottet N, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur Urol 2017;71:618-29. [Crossref] [PubMed]
- Louie KS, Seigneurin A, Cathcart P, et al. Do prostate cancer risk models improve the predictive accuracy of PSA screening? A meta-analysis. Ann Oncol 2015;26:848-64. [Crossref] [PubMed]
- van Buuren SG. Multivariate Imputation by Chained Equations in R. J Stat Softw 2011.45.
- Nieboer D, Vergouwe Y, Ankerst DP, et al. Improving prediction models with new markers: a comparison of updating strategies. BMC Med Res Methodol 2016;16:128. [Crossref] [PubMed]
- Chun FK, Steuber T, Erbersdobler A, et al. Development and internal validation of a nomogram predicting the probability of prostate cancer Gleason sum upgrading between biopsy and radical prostatectomy pathology. Eur Urol 2006;49:820-6. [Crossref] [PubMed]
- Roobol MJ, Steyerberg EW, Kranse R, et al. A risk-based strategy improves prostate-specific antigen-driven detection of prostate cancer. Eur Urol 2010;57:79-85. [Crossref] [PubMed]
- Finne P, Finne R, Bangma C, et al. Algorithms based on prostate-specific antigen (PSA), free PSA, digital rectal examination and prostate volume reduce false-positive PSA results in prostate cancer screening. Int J Cancer 2004;111:310-5. [Crossref] [PubMed]
- Karakiewicz PI, Benayoun S, Kattan MW, et al. Development and validation of a nomogram predicting the outcome of prostate biopsy based on patient age, digital rectal examination and serum prostate specific antigen. J Urol 2005;173:1930-4. [Crossref] [PubMed]
- Stephan C, Cammann H, Semjonow A, et al. Multicenter evaluation of an artificial neural network to increase the prostate cancer detection rate and reduce unnecessary biopsies. Clin Chem 2002;48:1279-87. [PubMed]
- Ankerst DP, Hoefler J, Bock S, et al. Prostate Cancer Prevention Trial risk calculator 2.0 for the prediction of low- vs high-grade prostate cancer. Urology 2014;83:1362-7. [Crossref] [PubMed]
- Nam RK, Toi A, Klotz LH, et al. Assessing individual risk for prostate cancer. J Clin Oncol 2007;25:3582-8. [Crossref] [PubMed]
- Roobol MJ, Verbeek JF, van der Kwast T, et al. Improving the rotterdam european randomized study of screening for prostate cancer risk calculator for initial prostate biopsy by incorporating the 2014 International Society of Urological Pathology Gleason Grading and Cribriform growth. Eur Urol 2017;72:45-51. [Crossref] [PubMed]
- Alberts AR, Schoots IG, Bokhorst LP, et al. Risk-based patient selection for magnetic resonance imaging-targeted prostate biopsy after negative transrectal ultrasound-guided random biopsy avoids unnecessary magnetic resonance imaging scans. Eur Urol 2016;69:1129-34. [Crossref] [PubMed]
- Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010;21:128-38. [Crossref] [PubMed]
- van Houwelingen HC, Arends LR, Stijnen T. Advanced methods in meta-analysis: multivariate approach and meta-regression. Stat Med 2002;21:589-624. [Crossref] [PubMed]
- Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 2016;352:i6. [Crossref] [PubMed]
- Kerr KF, Brown MD, Zhu K, et al. Assessing the clinical impact of risk prediction models with decision curves: guidance for correct interpretation and appropiate use. J Clin Oncol 2016;34:2534-40. [Crossref] [PubMed]
- Carlsson SV, de Carvalho TM, Roobol MJ, et al. Estimating the harms and benefits of prostate cancer screening as used in common practice versus recommended good practice: A microsimulation screening analysis. Cancer 2016;122:3386-93. [Crossref] [PubMed]
- Moyer VA, Force USPST. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2012;157:120-34. [Crossref] [PubMed]
- Bibbins-Domingo K, Grossman DC, Curry SJ. The US Preventive Services Task Force 2017 draft recommendation statement on screening for prostate cancer: an invitation to review and comment. JAMA 2017;317:1949-50. [Crossref] [PubMed]
- Parekh DJ, Punnen S, Sjoberg DD, et al. A multi-institutional prospective trial in the usa confirms that the 4kscore accurately identifies men with high-grade prostate cancer. Eur Urol 2015;68:464-70. [Crossref] [PubMed]
- Van Calster B, Nieboer D, Vergouwe Y, et al. A calibration hierarchy for risk models was defined: from utopia to empirical data. J Clin Epidemiol 2016;74:167-76. [Crossref] [PubMed]
- Schoots IG, Roobol MJ, Nieboer D, et al. Magnetic resonance imaging-targeted biopsy may enhance the diagnostic accuracy of significant prostate cancer detection compared to standard transrectal ultrasound-guided biopsy: a systematic review and meta-analysis. Eur Urol 2015;68:438-50. [Crossref] [PubMed]
- Roobol MJ, Vedder MM, Nieboer D, et al. Comparison of two prostate cancer risk calculators that include the prostate health index. Eur Urol Focus 2015;1:185-90. [Crossref] [PubMed]
- Vedder MM, de Bekker-Grob EW, Lilja HG, et al. The added value of percentage of free to total prostate-specific antigen, PCA3, and a kallikrein panel to the ERSPC risk calculator for prostate cancer in prescreened men. Eur Urol 2014;66:1109-15. [Crossref] [PubMed]
- Perdonà S, Cavadas V, Di Lorenzo G, et al. Prostate cancer detection in the "grey area" of prostate-specific antigen below 10 ng/ml: head-to-head comparison of the updated PCPT calculator and Chun's nomogram, two risk estimators incorporating prostate cancer antigen 3. Eur Urol 2011;59:81-7. [Crossref] [PubMed]
- Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol 1974;111:58-64. [Crossref] [PubMed]
- Loeb S, Folkvaljon Y, Robinson D, et al. Evaluation of the 2015 Gleason Grade Groups in a Nationwide Population-based Cohort. Eur Urol 2016;69:1135-41. [Crossref] [PubMed]
- Kweldam CF, Kümmerlin IP, Nieboer D, et al. Prostate cancer outcomes of men with biopsy Gleason score 6 and 7 without cribriform or intraductal carcinoma. Eur J Cancer 2016;66:26-33. [Crossref] [PubMed]


